26.4.11. B2BII¶
The b2bii package in basf2 converts Belle MDST files (basf data format) to
Belle II MDST (basf2 data format). This enables performing physics analysis
using data collected with Belle detector with the analysis software and
algorithms developed for the analysis of data collected with the Belle II
detector. It allows for estimation and validation of
performances of various advanced algorithms being developed for
Belle II
(such as Full Event Interpretation, Flavour Tagger, Tag Vertex, …).
B2BII converter¶
The b2bii convert reads and converts Belle MDST within basf2, and the converted data can be then analysed within the same job, without any intermediate steps.
The workflow is illustrated in this figure:
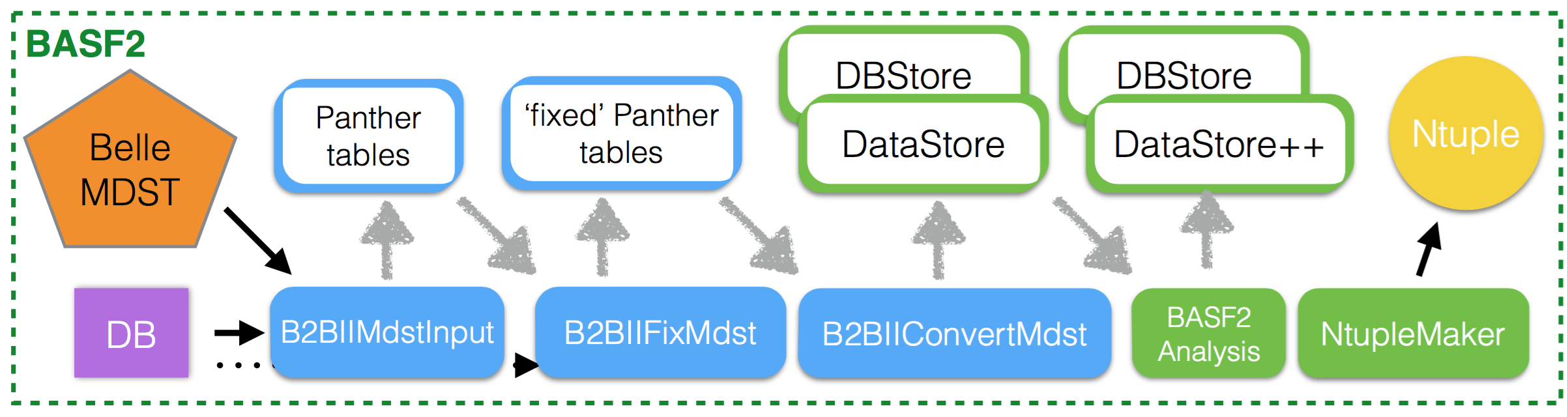
Read Belle MDST file.
Apply momentum or energy corrections in the objects.
Save objects in basf2 format
Time for basf2 analysis!
MC samples¶
As the Belle detector geometry is not implemented in basf2, one needs to either use the official Belle MC samples or generate own signal MC using basf. The information of generating Belle MC can be found here
Generic MC
Generic MC is the official MC samples at Belle, which was generated with
run-dependent beam background. There are multiple streams of these samples,
and each stream contains the same amount of events as present in the real
Belle data.
But which types of MC samples are exactly there? Similar to Belle II MC, there are also different categories of Belle MC:
Generic \(B\) samples : charged (\(B^+ B^-\)) and mixed (\(B^{0}\overline{B}^{0}\))
Continuum samples : uds, charm
Y(5S) samples : bsbs, nonbsbs
Generic \(B\) samples only contain decay modes with \(b \to c\) quark transitions, and have been generated based on the decay tables at
/sw/belle/belle/b20090127_0910/share/data-files/evtgenutil/DECAY.DEC.
How to find Generic MC samples?
You can find the sample(s) you want through Belle File Search Engine
Warning
Belle File Search Engine is only accessible within KEK domain or via VPN. Or look in the SSH - Secure Shell tutorial for a way to access it via SSH forwarding.
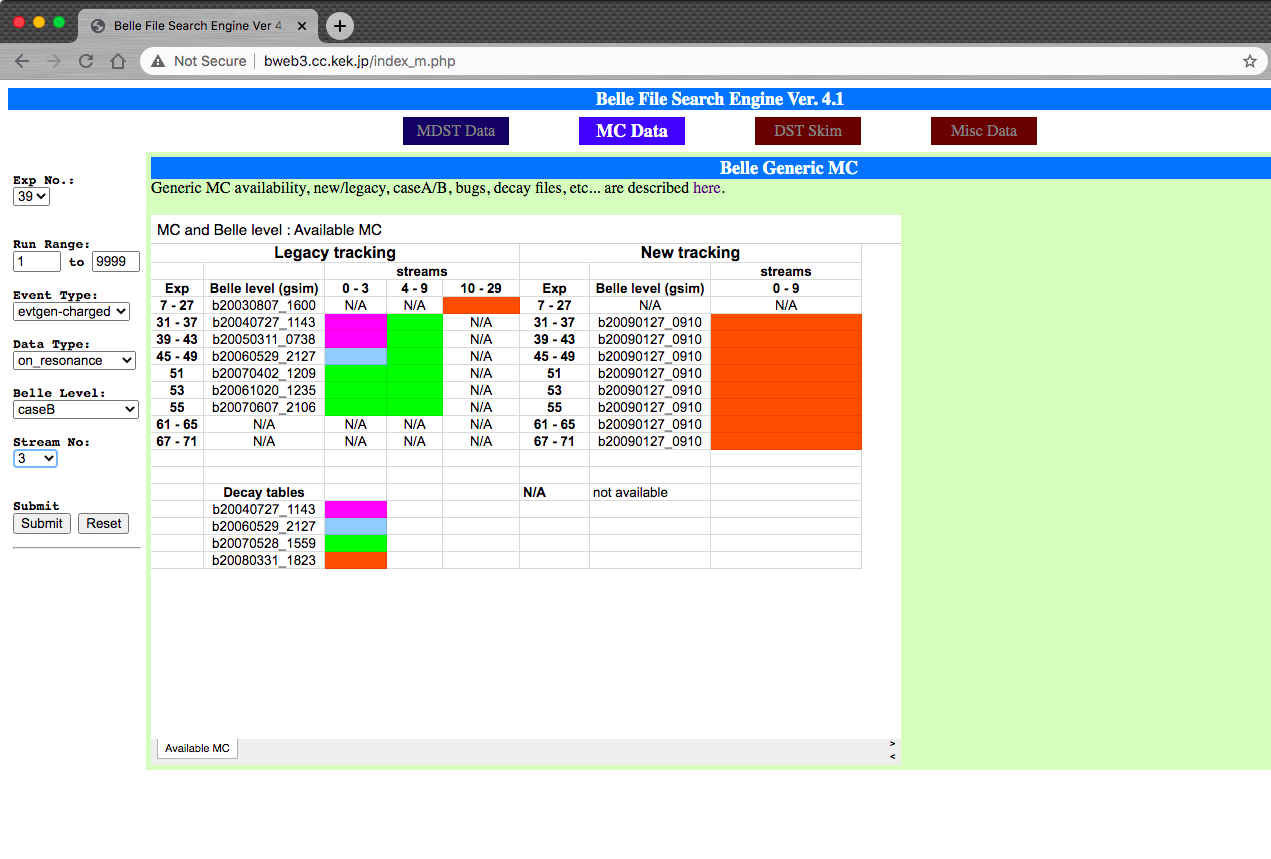
Fig. 26.28 The Belle File Search Engine¶
By specifying Exp No, Event Type, Data Type, and Stream No,
Event Type means different MC types (charged, mixed, uds, .. ).
Data Type is for different energy runs (on-resonance, off-resonance, …).
In total there are 10 streams of Generic \(B\) samples and 6 streams of
continuum samples.
You can either use the file list (physical path) or URL as input file list for b2bii jobs.
Note
Belle File Search Engine is also for data files.
See also
More information about official MC and data can be found here
Rare MC
Just from this name you can guess that this type of MC aims for rarer processes, such as \(b \to u \ell \nu\), \(e^+ e^- \to \tau^+ \tau^-\)…
Rare \(B\) MC was generated with an experiment-dependent beam energy, but not run-dependent (i.e. The same beam energy and IP profile in the same experiment).
The location of those special MC files can be found here.
Signal MC
As there is no Belle detector description, you can only use basf to produce signal MC samples.
Now we will learn how to use the mcproduzh package to generate signal MC in Belle.
This package was developed by “U”shiroda-san, A. “Z”upanc, and “H”orii-san, and
it consists of generation, simulation, and reconstruction based on evtgen and
gsim scripts (gsim is Belle slang for the simulated detector response
that results from the use of Geant3 within the Belle analysis software
framework basf – the output of gsim scripts are Belle mdst files).
It will create MC samples for a list of experiments, normalized by their \(N(B\overline{B})\) or integrated luminosity.
The beam energy, IP profile, and detector configuration of this MC will be experiment-dependent, but not run-dependent. Moreover, RunNo for these events will be set to 0, hence it doesn’t work for off-resonance or \(\Upsilon(nS)\).
First step: copy the file and unzip it
cp /home/belle/capid/public/B2SKW/mc/mcproduzh.tar.gz your_working_directory
tar -zxvf mcproduzh.tar.gz
There will be two directories evtgen and gsim, and one file READER.
Second step: generate events according to a decay table
Go to your evtgen directory
cd mcproduzh/evtgen
./runEvtgen nBB.txt [user-decay-table].dec [module-param-config].conf \
[TotalNomberOfEvents] [EventsPerJob]
[module-param-config].conf is for evtgen module configuration setting.
There are config setting examples in the package. For B analysis,
just choose Y4S.conf for your jobs.
In this step, you will get *.gen files stored in the
mcproduzh/evtgen/gen directory.
Finally, run the simulation and produce the mdst file. Go to gsim directory
cd mcproduzh/gsim/
./runGsimReco.csh "[absolutePathToEvtgenGeneratorFiles/]"
Warning
The path to the evtgen files has to be an absolute path!
Now you have MDST files produced in the mcproduzh/gsim/mdst/ directory.
Exercise
Try to generate a MC sample with 1000 \(B^{+} \to \overline{D}^{0}(\to K^{+} \pi^{-}) \pi^{+}\) events.
Solution
Generation:
cd <your_working_directory>/mcproduzh/evtgen
./runEvtgen nBB-Y4S.txt BptoD0pip-D0toKpi.dec Y4S.conf 1000 1000
Simulation:
cd ../gsim
./runGsimReco.csh <your_working_directory>/mcproduzh/evtgen/gen/
More information about MC can be found here.
Analysis with b2bii¶
With Belle MDST in hand, you can use it for your first b2bii analysis.
It is very simple, just replace inputMdst in your script with two
simple lines:
from b2biiConversion import convertBelleMdstToBelleIIMdst
convertBelleMdstToBelleIIMdst(inputfile, path=mypath)
Now we can use basf2 and analysis tools in basf2 to perform analyses over Belle MDST files.
The relations between basf and basf2 objects are shown in this figure:
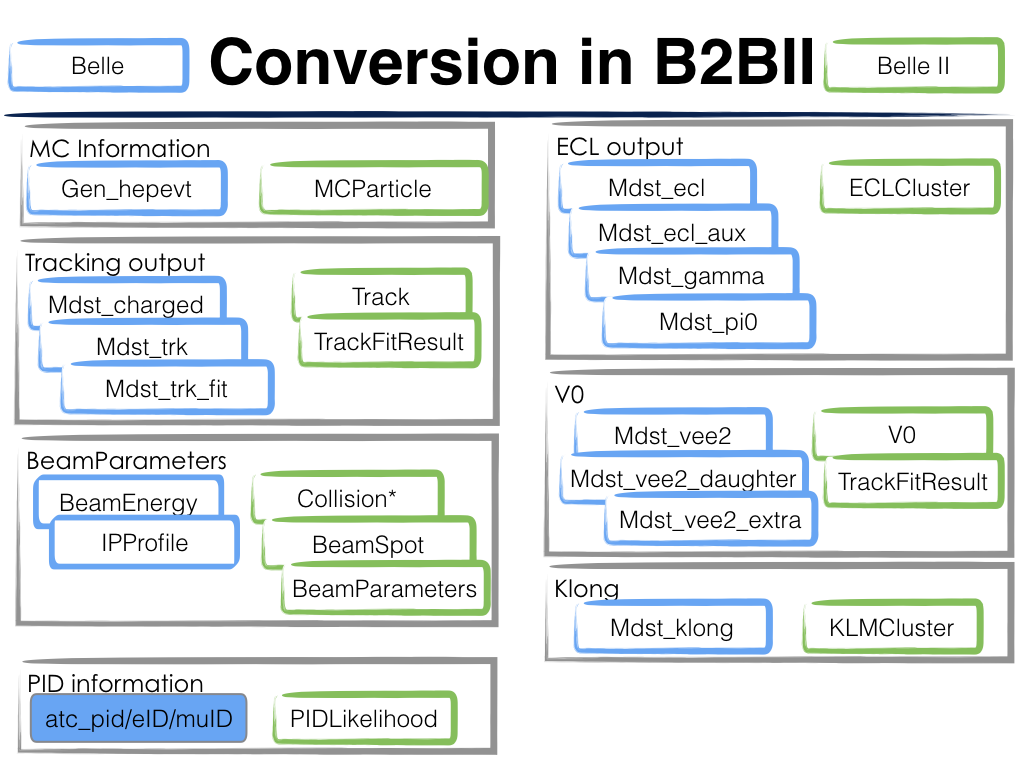
However, there are still many differences between the Belle detector and the Belle II detector, as well as basf and basf2. Therefore, we can’t simply use the same basf2 steering files, small modifications are needed.
Charged Final State Particles
basf and basf2 use different Helix parameterisations, however there exist a well
defined transformation from one parameterisation to the other.
Belle MDST stores in addition to the five helix parameters also the reference
point (or pivot point), which is assumed to be always point (0,0,0) in the
case of Belle II MDST.
Despite the different parameterisations, charged final state particles can still
be reconstructed using fillParticleList function in basf2.
But due to the different definition, as well as detector, it is not
recommended to use Belle II style PID in b2bii.
basf provided three different packages for PID:
atc_pid (KID) to separate kaons and pions, but also used for proton id
eID (electron ID) to separate electrons from hadrons
muid (muon ID) to separate muons from hadrons
Each of them in its own way combined information collected from various subdetector systems (CDC, ACC, TOF, ECL, KLM). The combination of individual likelihoods from each sub detector system is in some cases (eID) combined with the usage of external information, such as a priori probabilities of each particle type that is read from the Belle DB. Due to this fact the Belle-like PID probabilities can not be reproduced in basf2 from the raw likelihoods.
Alternatively, we can use the following predefined Belle-style PID variables to reproduce them:
Separation
basf2
Kaon vs pion
atcPIDBelle(3,2)
electron vs hadron
eIDBelle
muon likelihood
muIDBelle
muon likelihood quality flag
muIDBelleQuality
Exercise
Try to create a list of \(K^{+}\) with \(\text{KID}>0.6\), and another list of \(\pi^{+}\) with \(\text{KID}<0.4\).
Solution
import basf2
import modularAnalysis as ma
mypath = basf2.create_path()
ma.fillParticleList('K+:sig', 'atcPIDBelle(3,2)>0.6', path=mypath)
ma.fillParticleList('pi+:sig', 'atcPIDBelle(3,2)<0.4', path=mypath)
Neutral Final State Particles
Belle MDST has two additional data types: mdst_gamma and mdst_pi0,
for which there exist no equivalent data type in the Belle II MDST format.
In other words, photons and \(\pi^0\) particles are already created in basf.
During the conversion, b2bii converter by default creates gamma:mdst and pi0:mdst.
Warning
Don’t use fillParticleList to create photon candidates and
don’t reconstruct \(\pi^0\) candidates from pairs of two photons by yourself.
Exercise
Reconstruct the decay \(D^0 \to K^{-} \pi^{+} \pi^{0}\) with mass between 1.7 to 2.0 GeV in a b2bii analysis.
Hint
Always use premade particle lists for neutrals!
Solution
ma.reconstructDecay('D0:Kpipi0 -> K-:sig pi+:sig pi0:mdst', '1.7 < M < 2.0', path=mypath)
V0 Particles
As mentioned in Charged Final State Particles, all charged tracks are
parametrised with a helix with the reference point set to (0,0,0) in basf2.
This is not optimal in the case of V0s whose decay vertices can be far away
from the origin.
Therefore, all V0 candidates from the Mdst_Vee2 table in basf are converted to
Particles and saved in the particle lists K_S0:mdst, Lambda0:mdst,
and gamma:v0mdst.
The created particles have momentum and decay vertex position set to values
given in Belle’s Mdst_Vee2 table and their daughters particles with momentum
and position at the pivot equal to V0 decay vertex.
In addition, the quality indicators for \(K_{S}^{0}\) and
\(\Lambda^{0}\) can be used by simply calling goodBelleKshort and
goodBelleLambda, respectively.
Exercise
Select good Kshort from K_S0:mdst list.
Hint
Use cutAndCopyList to select candidates from an existing list.
Solution
ma.cutAndCopyList('K_S0:good', 'K_S0:mdst', cut='goodBelleKshort', path=mypath)
\(K_{L}^{0}\)
KLMClusters (Mdst_KLM_Cluster) and Klongs (Mdst_Klong) are converted.
The Klongs are stored in the default K_L0:mdst.
Warning
Don’t use fillParticleList to create Klong candidates.
Task
Use the final task in First steering file lesson as a template, try to convert it to your first b2bii analysis script.
This time, let’s reconstruct \(B^{-} \to D^{0} \pi^{-}\) with \(D^{0} \to K^{-}\pi^{+}\pi^{0}\).
Apply the same PID selections for your \(K\) and \(\pi\) as in the earlier exercise.
In the end, save the PID (Belle-style electronID, muonID, and KID), with other variables for all particles in the decay chain to the ntuple.
You can use this line to get the example file:
basf2.find_file('b2bii_input_evtgen_exp_07_BptoD0pip-D0toKpipi0-0.mdst', 'examples', False)
Hint
First steering file lesson is your best friend!
Remember always using premade particle lists for neutrals, Don’t forget to use Belle-style PID for charged particles.
Solution
1#!/usr/bin/env python3
2
3##########################################################################
4# basf2 (Belle II Analysis Software Framework) #
5# Author: The Belle II Collaboration #
6# #
7# See git log for contributors and copyright holders. #
8# This file is licensed under LGPL-3.0, see LICENSE.md. #
9##########################################################################
10
11import os
12
13import basf2 as b2
14import modularAnalysis as ma
15import variables as va
16import variables.collections as vc
17import variables.utils as vu
18from b2biiConversion import convertBelleMdstToBelleIIMdst
19
20os.environ["USE_GRAND_REPROCESS_DATA"] = "1"
21os.environ["PGUSER"] = "g0db"
22
23# Convert
24main = b2.create_path()
25inputfile = b2.find_file(
26 "b2bii_input_evtgen_exp_07_BptoD0pip-D0toKpipi0-0.mdst", "examples", False
27)
28convertBelleMdstToBelleIIMdst(inputfile, path=main)
29
30# Only charged final state particles need to be loaded. The neutral particles
31# gamma, pi0, K_S0, K_L0, and Lambda0 are already loaded to the 'gamma:mdst',
32# 'pi0:mdst', 'K_S0:mdst', 'K_L0:mdst', and 'Lambda0:mdst' particle lists,
33# respectively.
34ma.fillParticleList("pi+:sig", "atcPIDBelle(3,2)<0.4", path=main)
35ma.fillParticleList("K+:sig", "atcPIDBelle(3,2)>0.6", path=main)
36
37# The Belle PID variables are: atcPIDBelle(sigHyp, bkgHyp), muIDBelle, and eIDBelle
38va.variables.addAlias("Lkpi", "atcPIDBelle(3,2)")
39
40# Now, let's really reconstruct a B decay with an intermediate D meson:
41ma.reconstructDecay(
42 "D0:Kpipi0 -> K-:sig pi+:sig pi0:mdst", cut="1.7 < M < 2.0", path=main
43)
44ma.reconstructDecay(
45 "B+:D0pi -> anti-D0:Kpipi0 pi+:sig",
46 cut="4.8 < Mbc >5.2 and abs(deltaE) < 0.15",
47 path=main,
48)
49
50ma.matchMCTruth("B+:D0pi", path=main)
51
52# create and fill flat Ntuple with MCTruth and kinematic information
53# Create list of variables to save into the output file
54b_vars = []
55
56standard_vars = vc.kinematics + vc.mc_kinematics + vc.mc_truth + vc.inv_mass
57b_vars += vc.deltae_mbc
58b_vars += standard_vars
59
60# Variables for D0 and all final state particles (kaons, pions, neutral pions)
61b_vars += vu.create_aliases_for_selected(
62 standard_vars,
63 "B+ -> [ ^D0 -> ^K- ^pi+ ^pi0] ^pi+",
64 prefix=["D0", "K", "pip1", "pi0", "pip2"],
65)
66
67# Variables for charged final states (kaons, pions)
68belle1pid = (
69 ["eIDBelle", "muIDBelleQuality", "muIDBelle", "Lkpi"]
70 + vc.track
71 + vc.track_hits
72)
73
74b_vars += vu.create_aliases_for_selected(
75 belle1pid, "B+ -> [ D0 -> ^K- ^pi+ pi0] ^pi+", prefix=["K", "pip1", "pip2"]
76)
77
78ma.variablesToNtuple(
79 "B+:D0pi", b_vars, filename="B2BII_B2D0pi_D2Kpipi0.root", path=main
80)
81
82# progress
83main.add_module("Progress")
84
85b2.process(main)
86
87# Print call statistics
88print(b2.statistics)
Key points
Making basf2 process Belle data is as easy as adding
convertBelleMdstToBelle2Mdst()to the top of your steering file.Be careful with particle lists and variables in your analysis.
Never use
fillParticleListto create neutral final state particles!!
Stuck? We can help!
If you get stuck or have any questions to the online book material, the #starterkit-workshop channel in our chat is full of nice people who will provide fast help.
Refer to Collaborative Tools. for other places to get help if you have specific or detailed questions about your own analysis.
Improving things!
If you know how to do it, we recommend you to report bugs and other requests
with JIRA. Make sure to use the
documentation-training component of the Belle II Software project.
If you just want to give very quick feedback, use the last box “Quick feedback”.
Please make sure to be as precise as possible to make it easier for us to fix things! So for example:
typos (where?)
missing bits of information (what?)
bugs (what did you do? what goes wrong?)
too hard exercises (which one?)
etc.
If you are familiar with git and want to create your first pull request for the software, take a look at How to contribute. We’d be happy to have you on the team!
Quick feedback!
Author of this lesson
Chia-Ling Hsu