9.1.1. The Calibration Framework (CAF)¶
The Python CAF is a set of Python modules and classes which work together to provide users with a convienient interface to running calibration jobs. It is assumed that you are using the C++ calibration framework classes e.g. the CalibrationCollectorModule and CalibrationAlgorithm class in the calibration package. These provide a consistent interface for running calibration jobs on input data which the Python CAF can automate for you.
Essentially you can think of the Python CAF as providing a way to automatically create basf2 scripts that run your Collector module and Algorithm for you. That way you don’t have to create multiple shell submission scripts and any improvements made to the CAF will benefit you. It is important to remember that when you are running a CAF process, you are not running a basf2 process directly. You are instead running a Python script that will create basf2 processes for you.
The Calibration Class¶
The first part of the CAF to understand is the Calibration class.
This is where you specify which Collector module and which Algorithms you want to run.
It is also where you can configure the basf2 processes before they are run for you.
This includes setting the input data files, different global tags, and making reconstruction paths to run
prior to your Collector module.
-
class
caf.framework.Calibration(name, collector=None, algorithms=None, input_files=None, pre_collector_path=None, database_chain=None, output_patterns=None, max_files_per_collector_job=None, max_collector_jobs=None, backend_args=None)[source]¶ Bases:
caf.framework.CalibrationBaseEvery Calibration object must have at least one collector at least one algorithm. You have the option to add in your collector/algorithm by argument here, or add them later by changing the properties.
If you plan to use multiple
Collectionobjects I recommend that you only set the name here and add the Collections separately viaadd_collection().- Parameters
name (str) – Name of this calibration. It should be unique for use in the
CAF- Keyword Arguments
collector (str,
basf2.Module) – Should be set to a CalibrationCollectorModule() or a string with the module name.algorithms (list,
ROOT.Belle2.CalibrationAlgorithm) – The algorithm(s) to use for thisCalibration.input_files (str, list[str]) – Input files for use by this Calibration. May contain wildcards useable by
glob.glob
A Calibration won’t be valid in the
CAFuntil it has all of these four attributes set. For example:>>> cal = Calibration('TestCalibration1') >>> col1 = register_module('CaTest') >>> cal.add_collection('TestColl', col1)
or equivalently
>>> cal = Calibration('TestCalibration1', 'CaTest')
If you want to run a basf2
pathbefore your collector module when running over data>>> cal.pre_collector_path = my_basf2_path
You don’t have to put a RootInput module in this pre-collection path, but you can if you need some special parameters. If you want to process sroot files the you have to explicitly add SeqRootInput to your pre-collection path. The inputFileNames parameter of (Seq)RootInput will be set by the CAF automatically for you.
You can use optional arguments to pass in some/all during initialisation of the
Calibrationclass>>> cal = Calibration( 'TestCalibration1', 'CaTest', [alg1,alg2], ['/path/to/file.root'])
you can change the input file list later on, before running with
CAF>>> cal.input_files = ['path/to/*.root', 'other/path/to/file2.root']
If you have multiple collections from calling
add_collection()then you should instead set the pre_collector_path, input_files, database chain etc from there. SeeCollection.Adding the CalibrationAlgorithm(s) is easy
>>> alg1 = TestAlgo() >>> cal.algorithms = alg1
Or equivalently
>>> cal.algorithms = [alg1]
Or for multiple algorithms for one collector
>>> alg2 = TestAlgo() >>> cal.algorithms = [alg1, alg2]
Note that when you set the algorithms, they are automatically wrapped and stored as a Python class
Algorithm. To access the C++ algorithm clas underneath directly do:>>> cal.algorithms[i].algorithm
If you have a setup function that you want to run before each of the algorithms, set that with
>>> cal.pre_algorithms = my_function_object
If you want a different setup for each algorithm use a list with the same number of elements as your algorithm list.
>>> cal.pre_algorithms = [my_function1, my_function2, ...]
You can also specify the dependencies of the calibration on others
>>> cal.depends_on(cal2)
By doing this, the
CAFwill respect the ordering of the calibrations and will pass the calibration constants created by earlier completed calibrations to dependent ones.-
add_collection(name, collection)[source]¶ - Parameters
name (str) – Unique name of this
Collectionin the Calibration.collection (
Collection) –Collectionobject to use.
Adds a new
Collectionobject to theCalibration. Any valid Collection will be used in the Calibration. A default Collection is automatically added but isn’t valid and won’t run unless you have assigned a collector + input files. You can ignore the default one and only add your own custom Collections. You can configure the default from the Calibration(…) arguments or after creating the Calibration object via directly setting the cal.collector, cal.input_files attributes.
-
alg_output_dir= 'algorithm_output'¶ Subdirectory name for algorithm output
-
property
algorithms¶ Algorithmclasses that wil be run by thisCalibration. You should set this attribute to either a single CalibrationAlgorithm C++ class, or alistof them if you want to run multiple CalibrationAlgorithms using one CalibrationCollectorModule.
-
algorithms_runner= None¶ The class that runs all the algorithms in this Calibration using their assigned
caf.strategies.AlgorithmStrategy. Plugin your own runner class to change how your calibration will run the list of algorithms.
-
backend= None¶ The
backendwe’ll use for our collector submission in this calibration. IfNoneit will be set by the CAF used to run this Calibration (recommended!).
-
property
backend_args¶
-
checkpoint_states= ['init', 'collector_completed', 'completed']¶ Checkpoint states which we are allowed to restart from.
-
collections= None¶ Collections stored for this calibration.
-
property
collector¶
-
collector_full_update_interval= None¶ While checking if the collector is finished we don’t bother wastefully checking every subjob’s status. We exit once we find the first subjob that isn’t ready. But after this interval has elapsed we do a full
caf.backends.Job.update_status()call and print the fraction of SubJobs completed.
-
database_chain= None¶ The database chain that is applied to the algorithms. This is often updated at the same time as the database chain for the default
Collection.
-
default_collection_name= 'default'¶ Default collection name
-
property
files_to_iovs¶
-
heartbeat= None¶ This calibration’s sleep time before rechecking to see if it can move state
-
ignored_runs= None¶ List of ExpRun that will be ignored by this Calibration. This runs will not have Collector jobs run on them (if possible). And the algorithm execution will exclude them from a ExpRun list. However, the algorithm execution may merge IoVs of final payloads to cover the ‘gaps’ caused by these runs. You should pay attention to what the AlgorithmStrategy you choose will do in these cases.
-
property
input_files¶
-
is_valid()[source]¶ A full calibration consists of a collector AND an associated algorithm AND input_files.
- Returns False if:
We are missing any of the above.
There are multiple Collections and the Collectors have mis-matched granularities.
Any of our Collectors have granularities that don’t match what our Strategy can use.
-
property
iteration¶ Retrieves the current iteration number in the database file.
- Returns
The current iteration number
- Return type
-
machine= None¶ The
caf.state_machines.CalibrationMachinethat we will run to process this calibration start to finish.
-
property
max_collector_jobs¶
-
property
max_files_per_collector_job¶
-
max_iterations= None¶ Variable to define the maximum number of iterations for this calibration specifically. It overrides tha CAF calibration_defaults value if set.
-
moves= ['submit_collector', 'complete', 'run_algorithms', 'iterate', 'fail_fully']¶ Allowed transitions that we will use to progress
-
property
output_patterns¶
-
property
pre_algorithms¶ Callback run prior to each algorithm iteration.
-
property
pre_collector_path¶
-
reset_database(apply_to_default_collection=True)[source]¶ - Keyword Arguments
apply_to_default_collection (bool) – Should we also reset the default collection?
Remove everything in the database_chain of this Calibration, including the default central database tag automatically included from
basf2.conditions.default_globaltags. This will NOT affect the database chain of anyCollectionother than the default one. You can prevent the default Collection from having its chain reset by setting ‘apply_to_default_collection’ to False.
-
results= None¶ Output results of algorithms for each iteration
-
run()[source]¶ Main logic of the Calibration object. Will be run in a new Thread by calling the start() method.
-
property
state¶ The current major state of the calibration in the database file. The machine may have a different state.
-
property
strategies¶ The strategy that the algorithm(s) will be run against. Assign a list of strategies the same length as the number of algorithms, or assign a single strategy to apply it to all algorithms in this
Calibration. You can see the choices incaf.strategies.
-
use_central_database(global_tag, apply_to_default_collection=True)[source]¶ - Parameters
global_tag (str) – The central database global tag to use for this calibration.
- Keyword Arguments
apply_to_default_collection (bool) – Should we also call use_central_database on the default collection (if it exists)
Using this allows you to append a central database to the database chain for this calibration. The default database chain is just the central one from
basf2.conditions.default_globaltags. To turn off central database completely or use a custom tag as the base, you should callCalibration.reset_databaseand start adding databases withCalibration.use_local_databaseandCalibration.use_central_database.Note that the database chain attached to the
Calibrationwill only affect the defaultCollection(if it exists), and the algorithm processes. So calling:>> cal.use_central_database(“global_tag”)
will modify the database chain used by all the algorithms assigned to this
Calibration, and modifies the database chain assigned to>> cal.collections[‘default’].database_chain
But calling
>> cal.use_central_database(file_path, payload_dir, False)
will add the database to the Algorithm processes, but leave the default Collection database chain untouched. So if you have multiple Collections in this Calibration their database chains are separate. To specify an additional
CentralDatabasefor a different collection, you will have to call:>> cal.collections[‘OtherCollection’].use_central_database(“global_tag”)
-
use_local_database(filename, directory='', apply_to_default_collection=True)[source]¶ - Parameters
filename (str) – The path to the database.txt of the local database
- Keyword Argumemts:
directory (str): The path to the payloads directory for this local database. apply_to_default_collection (bool): Should we also call use_local_database on the default collection (if it exists)
Append a local database to the chain for this calibration. You can call this function multiple times and each database will be added to the chain IN ORDER. The databases are applied to this calibration ONLY. The Local and Central databases applied via these functions are applied to the algorithm processes and optionally the default
Collectionjob as a database chain. There are other databases applied to the processes later, checked by basf2 in this order:Local Database from previous iteration of this Calibration.
Local Database chain from output of previous dependent Calibrations.
This chain of Local and Central databases where the last added is checked first.
Note that this function on the
Calibrationobject will only affect the defaultCollectionif it exists and if ‘apply_to_default_collection’ remains True. So calling:>> cal.use_local_database(file_path, payload_dir)
will modify the database chain used by all the algorithms assigned to this
Calibration, and modifies the database chain assigned to>> cal.collections[‘default’].database_chain
But calling
>> cal.use_local_database(file_path, payload_dir, False)
will add the database to the Algorithm processes, but leave the default Collection database chain untouched.
If you have multiple Collections in this Calibration their database chains are separate. To specify an additional
LocalDatabasefor a different collection, you will have to call:>> cal.collections[‘OtherCollection’].use_local_database(file_path, payload_dir)
Overall the Calibration class basically runs a State Machine (SM).
Where it progresses from the initial state to completion or (failure) via several processing steps e.g. Submitting collector jobs.
Below is a simplified version of what is happening when caf.framework.Calibration.run() is called.
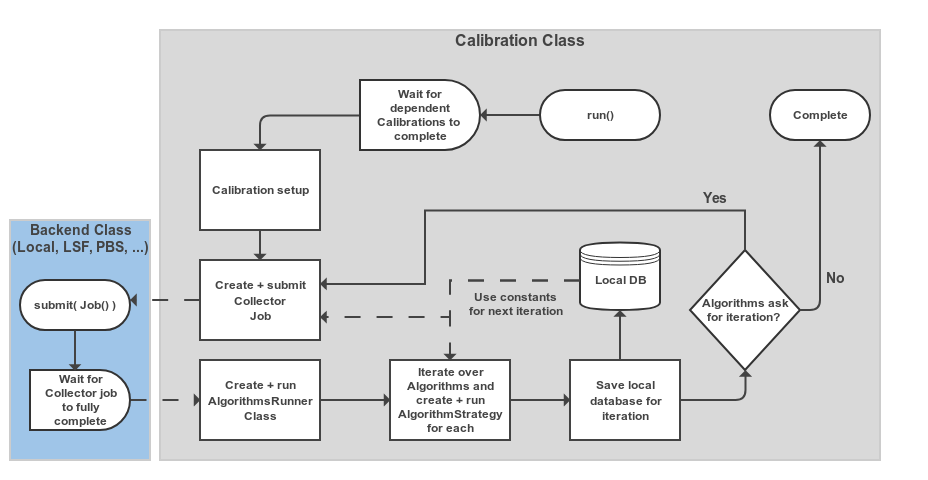
Fig. 9.1 Schematic view of the processing flow in a single Calibration object¶
The CAF Class¶
The caf.framework.CAF class is essentially an overall configuration object for the calibrations you want
to run in a single processing run.
You add Calibration objects to the CAF in order to have them run.
The CAF will start them when their dependent calibrations have completed.
The CAF also creates the overall output directory, checks the basic validity of calibrations (including cyclic-dependency)
before running, and assigns some default options to calibrations if they weren’t set in the calibration itself.
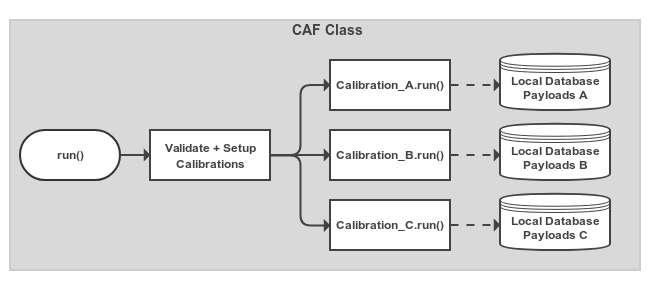
Fig. 9.2 Very simple view of the processing in the overall Calibration object.
Calibration objects that have been added are set up and have their caf.framework.Calibration.run() method called.
The Calibration threads started automatically wait until other Calibrations that they depend on have completed before
starting their main processing logic (see Fig. 9.1).¶
-
class
caf.framework.CAF(calibration_defaults=None)[source]¶ - Parameters
calibration_defaults (dict) –
A dictionary of default options for calibrations run by this
CAFinstance e.g.>>> calibration_defaults={"max_iterations":2}
This class holds
Calibrationobjects and processes them. It defines the initial configuration/setup for the calibrations. But most of the real processing is done through thecaf.state_machines.CalibrationMachine.The
CAFclass essentially does some initial setup, holds theCalibrationBaseinstances and calls theCalibrationBase.startwhen the dependencies are met.Much of the checking for consistency is done in this class so that no processing is done with an invalid setup. Choosing which files to use as input should be done from outside during the setup of the
CAFandCalibrationBaseinstances.-
add_calibration(calibration)[source]¶ Adds a
Calibrationthat is to be used in this program to the list. Also adds an empty dependency list to the overall dictionary. You should not directly alter aCalibrationobject after it has been added here.
-
property
backend¶ The
backendthat runs the collector job. When set, this is checked that abackends.Backendclass instance was passed in.
-
calibration_defaults= None¶ Default options applied to each calibration known to the
CAF, if theCalibrationhas these defined by the user then the defaults aren’t applied. A simple way to define the same configuration to all calibrations in theCAF.
-
calibrations= None¶ Dictionary of calibrations for this
CAFinstance. You should useadd_calibrationto add to this.
-
default_calibration_config= {'ignored_runs': [], 'max_iterations': 5}¶ The defaults for Calibrations
-
dependencies= None¶ Dictionary of dependencies of
Calibrationobjects, where value is the list ofCalibrationobjects that the key depends on. This attribute is filled during self.run()
-
future_dependencies= None¶ Dictionary of future dependencies of
Calibrationobjects, where the value is all calibrations that will depend on the key, filled during self.run()
-
heartbeat= None¶ The heartbeat (seconds) between polling for Calibrations that are finished
-
order= None¶ The ordering and explicit future dependencies of calibrations. Will be filled during
CAF.run()for you.
-
output_dir= None¶ Output path to store results of calibration and bookkeeping information
-
run(iov=None)[source]¶ - Keyword Arguments
iov (
caf.utils.IoV) – IoV to calibrate for this processing run. Only the input files necessary to calibrate this IoV will be used in the collection step.
This function runs the overall calibration job, saves the outputs to the output_dir directory, and creates database payloads.
Upload of final databases is not done here. This simply creates the local databases in the output directory. You should check the validity of your new local database before uploading to the conditions DB via the basf2 tools/interface to the DB.
The Algorithm Class¶
Warning
You probably don’t need to interact with this class unless you are attempting to set the
caf.framework.Algorithm.params dictionary.
-
class
caf.framework.Algorithm(algorithm, data_input=None, pre_algorithm=None)[source]¶ - Parameters
algorithm – The CalibrationAlgorithm instance that we want to execute.
- Keyword Arguments
data_input (types.FunctionType) – An optional function that sets the input files of the algorithm.
pre_algorithm (types.FunctionType) – An optional function that runs just prior to execution of the algorithm. Useful for set up e.g. module initialisation
This is a simple wrapper class around the C++ CalibrationAlgorithm class. It helps to add functionality to algorithms for use by the Calibration and CAF classes rather than separating the logic into those classes directly.
This is not currently a class that a user should interact with much during
CAFsetup (unless you’re doing something advanced). TheCalibrationclass should be doing the most of the creation of the defaults for these objects.Setting the
data_inputfunction might be necessary if you have set theCalibration.output_patterns. Also, setting thepre_algorithmto a function that should execute prior to eachstrategies.AlgorithmStrategyis often useful i.e. by calling for the Geometry module to initialise.-
algorithm= None¶ CalibrationAlgorithm instance (assumed to be true since the Calibration class checks)
-
data_input= None¶ Function called before the pre_algorithm method to setup the input data that the CalibrationAlgorithm uses. The list of files matching the
Calibration.output_patternsfrom the collector output directories will be passed to it
-
default_inputdata_setup(input_file_paths)[source]¶ Simple setup to set the input file names to the algorithm. Applied to the data_input attribute by default. This simply takes all files returned from the
Calibration.output_patternsand filters for only the CollectorOutput.root files. Then it sets them as input files to the CalibrationAlgorithm class.
-
name= None¶ The name of the algorithm, default is the Algorithm class name
-
params= None¶ Parameters that could be used in the execution of the algorithm strategy/runner to modify behaviour. By default this is empty and not used by the default
caf.strategies.SingleIOVclass. But more complex strategies, or your own custom ones, could use it to configure behaviour. Note that if you modify this inside a subprocess the modification won’t persist outside, you would have to change it in the parent process (or dump values to a file and read it in next time).
-
pre_algorithm= None¶ Function called after data_input but before algorithm.execute to do any remaining setup. It must have the form
pre_algorithm(algorithm, iteration)where algorithm can be assumed to be the CalibrationAlgorithm instance about to be executed, and iteration is an int e.g. 0, 1, 2…
-
strategy= None¶ The algorithm stratgey that will be used when running over the collected data. you can set this here, or from the
Calibration.strategiesattribute.
Restarting The CAF From Failure¶
During development you will likely find that sometimes your Calibration fails to complete due to problems in the code.
When using the full caf.framework.CAF class this can be quite problematic.
For example, if your algorithm fails after the collector step has already completed (which may take a long time depending
on how much data you’re using) then you might have to re-run your entire CAF process again.
In order to prevent this issue, the CAF uses a checkpoint system to save the state of each Calibration once it reaches a recoverable position in the workflow. If you run the CAF using the same output directory again the CAF will restart each Calibration requested from the checkpoint state of that Calibration i.e. the last recoverable state.
Put simply, if you get a failure when running the CAF try to fix the problem and then re-run your Python CAF script. The CAF should restart from a safe position and try to run the (now fixed) code again.
Multiple Collections¶
Sometimes you may have multiple data types which you want to use as input to your Calibration.
In this case you essentially want to run your Collector module with different pre-collection reconstruction
parameters and use all the data merged as input to the algorithm.
By using the Calibration.add_collection function you can add multiple different Collection objects to your
Calibration.
-
class
caf.framework.Collection(collector=None, input_files=None, pre_collector_path=None, database_chain=None, output_patterns=None, max_files_per_collector_job=None, max_collector_jobs=None, backend_args=None)[source]¶ - Keyword Arguments
collector (str, basf2.Module) – The collector module or module name for this
Collection.input_files (list[str]) – The input files to be used for only this
Collection.pre_collection_path (basf2.Path) – The reconstruction
basf2.Pathto be run prior to the Collector module.database_chain (list[CentralDatabase, LocalDatabase]) – The database chain to be used initially for this
Collection.output_patterns (list[str]) – Output patterns of files produced by collector which will be used to pass to the
Algorithm.data_inputfunction. Setting this here, replaces the default completely.max_files_for_collector_job (int) – Maximum number of input files sent to each collector subjob for this
Collection. Technically this sets the SubjobSplitter to be used, not compatible with max_collector_jobs.max_collector_jobs (int) – Maximum number of collector subjobs for this
Collection. Input files are split evenly between them. Technically this sets the SubjobSplitter to be used. Not compatible with max_files_for_collector_job.backend_args (dict) – The args for the backend submission of this
Collection.
-
backend_args= None¶ Dictionary passed to the collector Job object to configure how the
caf.backends.Backendinstance should treat the collector job when submitting. The choice of arguments here depends on which backend you plan on using.
-
property
collector¶ Collector module of this collection
-
database_chain= None¶ The database chain used for this Collection. NOT necessarily the same database chain used for the algorithm step! Since the algorithm will merge the collected data into one process it has to use a single DB chain set from the overall Calibration.
-
files_to_iovs= None¶ File -> Iov dictionary, should be
>>> {absolute_file_path:iov}
Where iov is a
IoVobject. Will be filled duringCAF.run()if empty. To improve performance you can fill this yourself before callingCAF.run()
-
property
input_files¶ Internal input_files stored for this calibration
-
job_cmd= None¶ The Collector
caf.backends.Job.cmdattribute. Probably using thejob_scriptto run basf2.
-
job_script= None¶ The basf2 steering file that will be used for Collector jobs run by this collection. This script will be copied into subjob directories as part of the input sandbox.
-
output_patterns= None¶ Output patterns of files produced by collector which will be used to pass to the
Algorithm.data_inputfunction. You can set these to anything understood byglob.glob, but if you want to specify this you should also specify theAlgorithm.data_inputfunction to handle the different types of files and call the CalibrationAlgorithm.setInputFiles() with the correct ones.
-
pre_collector_path= None¶ Since many collectors require some different setup, if you set this attribute to a
basf2.Pathit will be run before the collector and after the default RootInput module + HistoManager setup. If this path contains RootInput then it’s params are used in the RootInput module instead, except for the input_files parameter which is set to whichever files are passed to the collector subjob.
-
reset_database()[source]¶ Remove everything in the database_chain of this Calibration, including the default central database tag automatically included from
basf2.conditions.default_globaltags.
-
use_central_database(global_tag)[source]¶ - Parameters
global_tag (str) – The central database global tag to use for this calibration.
Using this allows you to add a central database to the head of the global tag database chain for this collection. The default database chain is just the central one from
basf2.conditions.default_globaltags. The input file global tag will always be overrided and never used unless explicitly set.To turn off central database completely or use a custom tag as the base, you should call
Calibration.reset_databaseand start adding databases withCalibration.use_local_databaseandCalibration.use_central_database.Alternatively you could set an empty list as the input database_chain when adding the Collection to the Calibration.
NOTE!! Since
release-04-00-00the behaviour of basf2 conditions databases has changed. All local database files MUST now be at the head of the ‘chain’, with all central database global tags in their own list which will be checked after all local database files have been checked.So even if you ask for
["global_tag1", "localdb/database.txt", "global_tag2"]to be the database chain, the real order that basf2 will use them is["global_tag1", "global_tag2", "localdb/database.txt"]where the file is checked first.
-
use_local_database(filename, directory='')[source]¶ - Parameters
Append a local database to the chain for this collection. You can call this function multiple times and each database will be added to the chain IN ORDER. The databases are applied to this collection ONLY.
NOTE!! Since release-04-00-00 the behaviour of basf2 conditions databases has changed. All local database files MUST now be at the head of the ‘chain’, with all central database global tags in their own list which will be checked after all local database files have been checked.
So even if you ask for [“global_tag1”, “localdb/database.txt”, “global_tag2”] to be the database chain, the real order that basf2 will use them is [“global_tag1”, “global_tag2”, “localdb/database.txt”] where the file is checked first.
Warning
If you are merging different data types in this way it is likely that they come from very different run ranges. Therefore your should take care that your AlgorithmStrategy setup makes sense and that you have checked that the output IoVs of the payloads are correct.
The b2caf-status Tool¶
In order to save these checkpoint states the CAF is creating a SQLite3 database file in your CAF.output_dir directory.
The b2caf-status command line tool lets you show the current status of the Calibrations even while the CAF is running.
It also lets you change the values in this database, so an advanced user could choose to reset a Calibration back to an earlier
iteration or checkpoint state.
This could be useful if a Collector step succeeded previously, but now needs to be re-run with different parameter values.
usage: b2caf-status [-h] [--path PATH] {show,update} ...
Optional Arguments
- --path, -p
The path to the CAF output directory you want to examine. By default it checks ‘./calibration_results’
Sub-commands:¶
show¶
Prints the current CAF database, showing values such as the current state, checkpoint, and iteration, for every Calibration known to the CAF process. You may call this even while the CAF is still running in order to check on the status.
b2caf-status show [-h]
update¶
This allows you to modify (update) the value of a column in the SQLite3 CAF database.
Do Not Do This While The CAF Is Running!
The CAF will automatically restart from whichever values ‘checkpoint’ and ‘iteration’ are set to in the DB the next time you run it. Therefore to restart a calibration from a specific point in the processing you should use this command to set the ‘checkpoint’ and ‘iteration’ columns to what you want. You can ignore the ‘state’ column e.g.
b2caf-status –path=my_output_dir update TestCal1 checkpoint init b2caf-status –path=my_output_dir update TestCal1 iteration 0
This will restart ‘TestCal1’ from the beginning completely, but will not affect any other Calibrations. Use the ‘b2caf-status show’ command to check that everything looks how you want it. The allowed values for the ‘checkpoint’ column are:
[‘init’, ‘collector_completed’, ‘completed’]
(Note that if you have developed your own Calibration class these allowed states may be different)
b2caf-status update [-h] cal_name col_name new_value
Required Arguments
- cal_name
Calibration name to update
- col_name
Column to change
- new_value
New value to update into the column
The b2caf-filemap Tool¶
Sometimes you will want to run over many input files. If you are ignoring certain runs from these files the CAF requires
that it knows which IoV each file corresponds to.
This is handled automatically by the CAF during startup, however this can take a long time to process if you have many files.
A better solution is to use the caf.framework.Calibration.files_to_iovs attribute and set a pre-calculated dictionary
manually.
To create this dictionary the b2caf-filemap tool can be used (though it isn’t necessary to use it) to create a pickle file
containing the dictionary.
Make a mapping file of file paths -> IoV. Outputs a dictionary in a pickle file to –output-file.
usage: b2caf-filemap [-h] [--method {raw,metadata}]
[--output-file OUTPUT_FILE] [--filter-regex FILTER_REGEX]
[--print-map]
filepath_pattern
Required Arguments
- filepath_pattern
The file path pattern that will be passed to glob. Selects the files to use in the map. e.g. “/hsm/belle2/bdata/Data/Raw/e0003/r0495[5,6]*/**/*.root”
Optional Arguments
- --method, -m
Possible choices: raw, metadata
The method by which you want to create the mapping. If your files are from ‘/hsm/belle2/bdata/Data/Raw’ then you can try to use ‘raw’. This is much faster as it generates the IoVs by using the directory structure of raw files. The ‘metadata’ option should work for all file paths but is slower as it has to use b2file-metadata-show to get the IoV of the files.
Default: “metadata”
- --output-file, -f
The output file that will contain the pickled file -> IoV map.
Default: “file_iov_map.pkl”
- --filter-regex, -re
A regular expression that will be applied to every filepath. Any filepath returning a value evaluating to True will be removed from the filepath list before the IoV map is generated. Use this to filter out specific files. We are using Python’s ‘re’ package with re.compile(‘regex’).search(filepath). e.g. Use ‘.bad’ to remove all filepaths containing the string ‘.bad’ anywhere in them.
- --print-map, -p
Prints the file -> IoV map to stdout as well.
Job Submission Backends¶
Quite often, your Collector processes will take some time to finish.
You may be collecting data from many files, with a large amount of pre-processing happening.
Running this all inside a single basf2 process is inefficient and could take days.
A better solution is to split the processing of the collector up into smaller processes that take a smaller
number of input files.
Without the CAF, basf2 users might create shell scripts and run them from the Python multiprocessing module.
Or they could submit to a Batch queue system on KEKCC using the bsub command.
To make this easier a small Python interface to job submission has been created in the backends module.
The CAF uses classes inheriting from Backend to submit the collector jobs to either
local multiprocessing, or several Batch queue systems like LSF used at KEKCC.
Although the CAF uses these classes, they are general enough that they can also be used from any Python program using the
basf2 library to submit commands/shell scripts.
You will first want to create a Job object to configure the process(es) you want to run.
-
class
caf.backends.Job(name, job_dict=None)[source]¶ This generic Job object is used to tell a Backend what to do. This object basically holds necessary information about a process you want to submit to a
Backend. It should not do anything that is backend specific, just hold the configuration for a job to be successfully submitted and monitored using a backend. The result attribute is where backend specific job monitoring goes.- Parameters
name (str) – Simply a name to describe the Job, not used for any critical purpose in the CAF
Warning
It is recommended to always use absolute paths for files when submitting a
Job.-
append_current_basf2_setup_cmds()[source]¶ This adds simple setup commands like
source /path/to/tools/b2setupto yourJob. It should detect if you are using a local release or CVMFS and append the correct commands so that the job will have the same basf2 release environment. It should also detect if a local release is not compiled with theoptoption.Note that this doesn’t mean that every environment variable is inherited from the submitting process environment.
-
args= None¶ The arguments that will be applied to the
cmd(These are ignored by SubJobs as they have their own arguments)
-
backend_args= None¶ Config dictionary for the backend to use when submitting the job. Saves us from having multiple attributes that may or may not be used.
-
check_input_data_files()[source]¶ Check the input files and make sure that there aren’t any duplicates. Also check if the files actually exist if possible.
-
cmd= None¶ Command and arguments as a list that wil be run by the job on the backend
-
copy_input_sandbox_files_to_working_dir()[source]¶ Get all of the requested files for the input sandbox and copy them to the working directory. Files like the submit.sh or input_data.json are not part of this process.
-
create_subjob(i, input_files=None, args=None)[source]¶ Creates a subjob Job object that references that parent Job. Returns the SubJob object at the end.
-
dump_input_data()[source]¶ Dumps the
Job.input_filesattribute to a JSON file. input_files should be a list of string URI objects.
-
dump_to_json(file_path)[source]¶ Dumps the Job object configuration to a JSON file so that it can be read in again later.
- Parameters
file_path (
Path) – The filepath we’ll dump to
-
exit_statuses= ['failed', 'completed']¶ Job statuses that correspond to the Job being finished (successfully or not)
-
property
full_command¶ Returns: str: The full command that this job will run including any arguments.
-
property
job_dict¶ Returns: dict: A JSON serialisable representation of the
Joband itsSubJobobjects.Pathobjects are converted to string viaPath.as_posix().
-
name= None¶ Job object’s name. Only descriptive, not necessarily unique.
-
property
output_dir¶ Output directory (
pathlib.Path), where we will download our output_files to. Default is ‘.’
-
output_patterns= None¶ Files that we produce during the job and want to be returned. Can use wildcard (*)
-
ready()[source]¶ Returns whether or not the Job has finished. If the job has subjobs then it will return true when they are all finished. It will return False as soon as it hits the first failure. Meaning that you cannot guarantee that all subjobs will have their status updated when calling this method. Instead use
update_status()to update all statuses if necessary.
-
result= None¶ The result object of this Job. Only filled once the job is submitted to a backend since the backend creates a special result class depending on its type.
-
setup_cmds= None¶ Bash commands to run before the main self.cmd (mainly used for batch system setup)
-
splitter= None¶ The
SubjobSplitterused to create subjobs if necessary
-
property
status¶ Returns the status of this Job. If the job has subjobs then it will return the overall status equal to the lowest subjob status in the hierarchy of statuses in
Job.statuses.
-
statuses= {'completed': 4, 'failed': 3, 'init': 0, 'running': 2, 'submitted': 1}¶ Allowed Job status dictionary. The key is the status name and the value is its level. The lowest level out of all subjobs is the one that is the overal status of the overall job.
-
subjobs= None¶ dict of subjobs assigned to this job
-
update_status()[source]¶ Calls
update_status()on the job’s result. The result object should update all of the subjobs (if there are any) in the best way for the type of result object/backend.
-
property
working_dir¶ Working directory of the job (
pathlib.Path). Default is ‘.’, mostly used in Local() backend
After creating a Job and configuring it, you need to actually run it.
The backends module provides several classes inheiriting from the
Backend class which you can use to submit a Job.
-
class
caf.backends.Backend(*, backend_args=None)[source]¶ Abstract base class for a valid backend. Classes derived from this will implement their own submission of basf2 jobs to whatever backend they describe. Some common methods/attributes go into this base class.
For backend_args the priority from lowest to highest is:
backend.default_backend_args -> backend.backend_args -> job.backend_args
-
backend_args= None¶ The backend args that will be applied to jobs unless the job specifies them itself
-
default_backend_args= {}¶ Default backend_args
-
exit_code_file= '__BACKEND_CMD_EXIT_STATUS__'¶ Default exit code file name
-
get_submit_script_path(job)[source]¶ Construct the Path object of the bash script file that we will submit. It will contain the actual job command, wrapper commands, setup commands, and any batch directives
-
abstract
submit(job)[source]¶ Base method for submitting collection jobs to the backend type. This MUST be implemented for a correctly written backend class deriving from Backend().
-
submit_script= 'submit.sh'¶ Default submission script name
-
The most simple option that should work everywhere is the Local backend.
This runs the Job using a Pool object with a configurable process Pool size.
-
class
caf.backends.Local(*, backend_args=None, max_processes=1)[source]¶ Backend for local processes i.e. on the same machine but in a subprocess.
Note that you should call the self.join() method to close the pool and wait for any running processes to finish before exiting the process. Once you’ve called join you will have to set up a new instance of this backend to create a new pool. If you don’t call
Local.joinor don’t create a join yourself somewhere, then the main python process might end before your pool is done.- Keyword Arguments
max_processes (int) – Integer that specifies the size of the process pool that spawns the subjobs, default=1. It’s the maximium simultaneous subjobs. Try not to specify a large number or a number larger than the number of cores. It won’t crash the program but it will slow down and negatively impact performance.
-
class
LocalResult(job, result)[source]¶ Result class to help monitor status of jobs submitted by Local backend.
-
get_exit_code_from_file()¶ Read the exit code file to discover the exit status of the job command. Useful falback if the job is no longer known to the job database (batch system purged it for example). Since some backends may take time to download the output files of the job back to the working directory we use a time limit on how long to wait.
-
ready()¶ Returns whether or not this job result is known to be ready. Doesn’t actually change the job status. Just changes the ‘readiness’ based on the known job status.
-
result= None¶ The underlying result from the backend
-
-
get_submit_script_path(job)¶ Construct the Path object of the bash script file that we will submit. It will contain the actual job command, wrapper commands, setup commands, and any batch directives
-
join()[source]¶ Closes and joins the Pool, letting you wait for all results currently still processing.
-
property
max_processes¶ The size of the multiprocessing process pool.
-
pool= None¶ The actual
Poolobject of this instance of the Backend.
If you have dozens of input files containing thousands of events, it may be slow to run your processing using the
Local backend backend.
At this point you should consider using a backend inheriting from the Batch backend class.
-
class
caf.backends.Batch(*, backend_args=None)[source]¶ Abstract Base backend for submitting to a local batch system. Batch system specific commands should be implemented in a derived class. Do not use this class directly!
-
can_submit(*args, **kwargs)[source]¶ Should be implemented in a derived class to check that submitting the next job(s) shouldn’t fail. This is initially meant to make sure that we don’t go over the global limits of jobs (submitted + running).
- Returns
If the job submission can continue based on the current situation.
- Return type
-
default_sleep_between_submission_checks= 30¶ Default time betweeon re-checking if the active jobs is below the global job limit.
-
get_batch_submit_script_path(job)[source]¶ Construct the Path object of the script file that we will submit using the batch command. For most batch backends this is the same script as the bash script we submit. But for some they require a separate submission file that describes the job. To implement that you can implement this function in the Backend class.
-
get_submit_script_path(job)¶ Construct the Path object of the bash script file that we will submit. It will contain the actual job command, wrapper commands, setup commands, and any batch directives
-
submission_cmds= []¶ Shell command to submit a script, should be implemented in the derived class
-
Currently there are three Batch classes.
The LSF, the PBS, and the
HTCondor backends.
If you are running at KEKCC (or any site providing the bsub command) you should use the LSF
backend.
If you are running at a site that uses the qsub command, you should use the PBS backend.
For NAF (DESY) and BNL you should use the HTCondor backends (they use condor_submit).
-
class
caf.backends.LSF(*, backend_args=None)[source]¶ Backend for submitting calibration processes to a qsub batch system.
-
class
LSFResult(job, job_id)[source]¶ Simple class to help monitor status of jobs submitted by LSF Backend.
You pass in a
Jobobject and job id from a bsub command. When you call thereadymethod it runs bjobs to see whether or not the job has finished.-
backend_code_to_status= {'DONE': 'completed', 'EXIT': 'failed', 'FINISHED': 'completed', 'PEND': 'submitted', 'RUN': 'running'}¶ LSF statuses mapped to Job statuses
-
get_exit_code_from_file()¶ Read the exit code file to discover the exit status of the job command. Useful falback if the job is no longer known to the job database (batch system purged it for example). Since some backends may take time to download the output files of the job back to the working directory we use a time limit on how long to wait.
-
job_id= None¶ job id given by LSF
-
ready()¶ Returns whether or not this job result is known to be ready. Doesn’t actually change the job status. Just changes the ‘readiness’ based on the known job status.
-
-
classmethod
bjobs(output_fields=None, job_id='', username='', queue='')[source]¶ Simplistic interface to the
bjobscommand. lets you request information about all jobs matching the filters ‘job_id’, ‘username’, and ‘queue’. The result is the JSON dictionary returned by output of the-jsonbjobs option.- Parameters
output_fields (list[str]) – A list of bjobs -o fields that you would like information about e.g. [‘stat’, ‘name’, ‘id’]
job_id (str) – String representation of the Job ID given by bsub during submission If this argument is given then the output of this function will be only information about this job. If this argument is not given, then all jobs matching the other filters will be returned.
username (str) – By default bjobs (and this function) return information about only the current user’s jobs. By giving a username you can access the job information of a specific user’s jobs. By giving
username='all'you will receive job information from all known user jobs matching the other filters.queue (str) – Set this argument to receive job information about jobs that are in the given queue and no other.
- Returns
JSON dictionary of the form:
{ "NJOBS":<njobs returned by command>, "JOBS":[ { <output field: value>, ... }, ... ] }
- Return type
-
classmethod
bqueues(output_fields=None, queues=None)[source]¶ Simplistic interface to the
bqueuescommand. lets you request information about all queues matching the filters. The result is the JSON dictionary returned by output of the-jsonbqueues option.- Parameters
output_fields (list[str]) – A list of bqueues -o fields that you would like information about e.g. the default is [‘queue_name’ ‘status’ ‘max’ ‘njobs’ ‘pend’ ‘run’]
queues (list[str]) – Set this argument to receive information about only the queues that are requested and no others. By default you will receive information about all queues.
- Returns
JSON dictionary of the form:
{ "COMMAND":"bqueues", "QUEUES":46, "RECORDS":[ { "QUEUE_NAME":"b2_beast", "STATUS":"Open:Active", "MAX":"200", "NJOBS":"0", "PEND":"0", "RUN":"0" }, ... }
- Return type
-
can_submit(njobs=1)[source]¶ Checks the global number of jobs in LSF right now (submitted or running) for this user. Returns True if the number is lower that the limit, False if it is higher.
- Parameters
njobs (int) – The number of jobs that we want to submit before checking again. Lets us check if we are sufficiently below the limit in order to (somewhat) safely submit. It is slightly dangerous to assume that it is safe to submit too many jobs since there might be other processes also submitting jobs. So njobs really shouldn’t be abused when you might be getting close to the limit i.e. keep it <=250 and check again before submitting more.
-
cmd_name= '#BSUB -J'¶ Job name directive
-
cmd_queue= '#BSUB -q'¶ Queue directive
-
cmd_stderr= '#BSUB -e'¶ stderr file directive
-
cmd_stdout= '#BSUB -o'¶ stdout file directive
-
cmd_wkdir= '#BSUB -cwd'¶ Working directory directive
-
default_backend_args= {'queue': 's'}¶ Default backend args for LSF
-
default_global_job_limit= 15000¶ Default global limit on the number of jobs to have in the system at any one time.
-
get_batch_submit_script_path(job)¶ Construct the Path object of the script file that we will submit using the batch command. For most batch backends this is the same script as the bash script we submit. But for some they require a separate submission file that describes the job. To implement that you can implement this function in the Backend class.
-
get_submit_script_path(job)¶ Construct the Path object of the bash script file that we will submit. It will contain the actual job command, wrapper commands, setup commands, and any batch directives
-
submission_cmds= ['bsub', '-env', '"none"', '<']¶ Shell command to submit a script
-
submit(job, check_can_submit=True, jobs_per_check=100)¶
-
class
-
class
caf.backends.PBS(*, backend_args=None)[source]¶ Backend for submitting calibration processes to a qsub batch system.
-
class
PBSResult(job, job_id)[source]¶ Simple class to help monitor status of jobs submitted by
PBSBackend.You pass in a
Jobobject (orSubJob) and job id from a qsub command. When you call thereadymethod it runs bjobs to see whether or not the job has finished.-
backend_code_to_status= {'C': 'completed', 'E': 'failed', 'FINISHED': 'completed', 'H': 'submitted', 'Q': 'submitted', 'R': 'running', 'T': 'submitted', 'W': 'submitted'}¶ PBS statuses mapped to Job statuses
-
get_exit_code_from_file()¶ Read the exit code file to discover the exit status of the job command. Useful falback if the job is no longer known to the job database (batch system purged it for example). Since some backends may take time to download the output files of the job back to the working directory we use a time limit on how long to wait.
-
job_id= None¶ job id given by PBS
-
ready()¶ Returns whether or not this job result is known to be ready. Doesn’t actually change the job status. Just changes the ‘readiness’ based on the known job status.
-
-
can_submit(njobs=1)[source]¶ Checks the global number of jobs in PBS right now (submitted or running) for this user. Returns True if the number is lower that the limit, False if it is higher.
- Parameters
njobs (int) – The number of jobs that we want to submit before checking again. Lets us check if we are sufficiently below the limit in order to (somewhat) safely submit. It is slightly dangerous to assume that it is safe to submit too many jobs since there might be other processes also submitting jobs. So njobs really shouldn’t be abused when you might be getting close to the limit i.e. keep it <=250 and check again before submitting more.
-
cmd_name= '#PBS -N'¶ Job name directive
-
cmd_queue= '#PBS -q'¶ Queue directive
-
cmd_stderr= '#PBS -e'¶ stderr file directive
-
cmd_stdout= '#PBS -o'¶ stdout file directive
-
cmd_wkdir= '#PBS -d'¶ Working directory directive
-
static
create_job_record_from_element(job_elem)[source]¶ Creates a Job dictionary with various job information from the XML element returned by qstat.
- Parameters
job_elem (xml.etree.ElementTree.Element) – The XML Element of the Job
- Returns
JSON serialisable dictionary of the Job information we are interested in.
- Return type
-
default_backend_args= {'queue': 'short'}¶ Default backend_args for PBS
-
default_global_job_limit= 5000¶ Default global limit on the number of jobs to have in the system at any one time.
-
get_batch_submit_script_path(job)¶ Construct the Path object of the script file that we will submit using the batch command. For most batch backends this is the same script as the bash script we submit. But for some they require a separate submission file that describes the job. To implement that you can implement this function in the Backend class.
-
get_submit_script_path(job)¶ Construct the Path object of the bash script file that we will submit. It will contain the actual job command, wrapper commands, setup commands, and any batch directives
-
classmethod
qstat(username='', job_ids=None)[source]¶ Simplistic interface to the
qstatcommand. Lets you request information about all jobs or ones matching the filter [‘job_id’] or for the username. The result is a JSON dictionary containing come of the useful job attributes returned by qstat.PBS is kind of annoying as depending on the configuration it can forget about jobs immediately. So the status of a finished job is VERY hard to get. There are other commands that are sometimes included that may do a better job. This one should work for Melbourne’s cloud computing centre.
- Keyword Arguments
username (str) – The username of the jobs we are interested in. Only jobs corresponding to the <username>@hostnames will be in the output dictionary.
job_ids (list[str]) – List of Job ID strings, each given by qstat during submission. If this argument is given then the output of this function will be only information about this jobs. If this argument is not given, then all jobs matching the other filters will be returned.
- Returns
JSON dictionary of the form (to save you parsing the XML that qstat returns).:
{ "NJOBS": int "JOBS":[ { <key: value>, ... }, ... ] }
- Return type
-
submission_cmds= ['qsub']¶ Shell command to submit a script
-
submit(job, check_can_submit=True, jobs_per_check=100)¶
-
class
-
class
caf.backends.HTCondor(*, backend_args=None)[source]¶ Backend for submitting calibration processes to a HTCondor batch system.
-
class
HTCondorResult(job, job_id)[source]¶ Simple class to help monitor status of jobs submitted by HTCondor Backend.
You pass in a
Jobobject and job id from a condor_submit command. When you call thereadymethod it runs condor_q and, if needed,condor_historyto see whether or not the job has finished.-
backend_code_to_status= {0: 'submitted', 1: 'submitted', 2: 'running', 3: 'failed', 4: 'completed', 5: 'submitted', 6: 'failed'}¶ HTCondor statuses mapped to Job statuses
-
get_exit_code_from_file()¶ Read the exit code file to discover the exit status of the job command. Useful falback if the job is no longer known to the job database (batch system purged it for example). Since some backends may take time to download the output files of the job back to the working directory we use a time limit on how long to wait.
-
job_id= None¶ job id given by HTCondor
-
ready()¶ Returns whether or not this job result is known to be ready. Doesn’t actually change the job status. Just changes the ‘readiness’ based on the known job status.
-
-
batch_submit_script= 'submit.sub'¶ HTCondor batch script (different to the wrapper script of
Backend.submit_script)
-
can_submit(njobs=1)[source]¶ Checks the global number of jobs in HTCondor right now (submitted or running) for this user. Returns True if the number is lower that the limit, False if it is higher.
- Parameters
njobs (int) – The number of jobs that we want to submit before checking again. Lets us check if we are sufficiently below the limit in order to (somewhat) safely submit. It is slightly dangerous to assume that it is safe to submit too many jobs since there might be other processes also submitting jobs. So njobs really shouldn’t be abused when you might be getting close to the limit i.e. keep it <=250 and check again before submitting more.
-
classmethod
condor_history(class_ads=None, job_id='', username='')[source]¶ Simplistic interface to the
condor_historycommand. lets you request information about all jobs matching the filtersjob_idandusername. Note that setting job_id negates username so it is ignored. The result is a JSON dictionary filled by output of the-jsoncondor_historyoption.- Parameters
class_ads (list[str]) – A list of condor_history ClassAds that you would like information about. By default we give {cls.default_class_ads}, increasing the amount of class_ads increase the time taken by the condor_q call.
job_id (str) – String representation of the Job ID given by condor_submit during submission. If this argument is given then the output of this function will be only information about this job. If this argument is not given, then all jobs matching the other filters will be returned.
username (str) – By default we return information about only the current user’s jobs. By giving a username you can access the job information of a specific user’s jobs. By giving
username='all'you will receive job information from all known user jobs matching the other filters. This is limited to 10000 records and isn’t recommended.
- Returns
JSON dictionary of the form:
{ "NJOBS":<number of records returned by command>, "JOBS":[ { <ClassAd: value>, ... }, ... ] }
- Return type
-
classmethod
condor_q(class_ads=None, job_id='', username='')[source]¶ Simplistic interface to the
condor_qcommand. lets you request information about all jobs matching the filters ‘job_id’ and ‘username’. Note that setting job_id negates username so it is ignored. The result is the JSON dictionary returned by output of the-jsoncondor_q option.- Parameters
class_ads (list[str]) – A list of condor_q ClassAds that you would like information about. By default we give {cls.default_class_ads}, increasing the amount of class_ads increase the time taken by the condor_q call.
job_id (str) – String representation of the Job ID given by condor_submit during submission. If this argument is given then the output of this function will be only information about this job. If this argument is not given, then all jobs matching the other filters will be returned.
username (str) – By default we return information about only the current user’s jobs. By giving a username you can access the job information of a specific user’s jobs. By giving
username='all'you will receive job information from all known user jobs matching the other filters. This may be a LOT of jobs so it isn’t recommended.
- Returns
JSON dictionary of the form:
{ "NJOBS":<number of records returned by command>, "JOBS":[ { <ClassAd: value>, ... }, ... ] }
- Return type
-
default_backend_args= {'extra_lines': [], 'getenv': 'false', 'path_prefix': '', 'request_memory': '4 GB', 'universe': 'vanilla'}¶ Default backend args for HTCondor
-
default_class_ads= ['GlobalJobId', 'JobStatus', 'Owner']¶ Default ClassAd attributes to return from commands like condor_q
-
default_global_job_limit= 10000¶ Default global limit on the number of jobs to have in the system at any one time.
-
get_batch_submit_script_path(job)[source]¶ Construct the Path object of the .sub file that we will use to describe the job.
-
get_submit_script_path(job)¶ Construct the Path object of the bash script file that we will submit. It will contain the actual job command, wrapper commands, setup commands, and any batch directives
-
submission_cmds= ['condor_submit', '-terse']¶ Batch submission commands for HTCondor
-
submit(job, check_can_submit=True, jobs_per_check=100)¶
-
class
Configuring SubJob Creation¶
You probably want to use basically the same setup for many (possibly 1000’s) of jobs but with different input data or
arguments. For this situation you can create SubJob objects of Job objects. SubJobs inherit most of the same attributes
as the parent job. However they can have different input files and arguments.
-
class
caf.backends.SubJob(job, subjob_id, input_files=None)[source]¶ This mini-class simply holds basic information about which subjob you are and a reference to the parent Job object to be able to access the main data there. Rather than replicating all of the parent job’s configuration again.
-
append_current_basf2_setup_cmds()¶ This adds simple setup commands like
source /path/to/tools/b2setupto yourJob. It should detect if you are using a local release or CVMFS and append the correct commands so that the job will have the same basf2 release environment. It should also detect if a local release is not compiled with theoptoption.Note that this doesn’t mean that every environment variable is inherited from the submitting process environment.
-
args= None¶ Arguments specific to this SubJob
-
check_input_data_files()¶ Check the input files and make sure that there aren’t any duplicates. Also check if the files actually exist if possible.
-
copy_input_sandbox_files_to_working_dir()¶ Get all of the requested files for the input sandbox and copy them to the working directory. Files like the submit.sh or input_data.json are not part of this process.
-
create_subjob(i, input_files=None, args=None)¶ Creates a subjob Job object that references that parent Job. Returns the SubJob object at the end.
-
dump_input_data()¶ Dumps the
Job.input_filesattribute to a JSON file. input_files should be a list of string URI objects.
-
dump_to_json(file_path)¶ Dumps the Job object configuration to a JSON file so that it can be read in again later.
- Parameters
file_path (
Path) – The filepath we’ll dump to
-
property
full_command¶ Returns: str: The full command that this job will run including any arguments.
-
id= None¶ Id of Subjob
-
property
job_dict¶ Returns: dict: A JSON serialisable representation of the
SubJob.Pathobjects are converted tostringviaPath.as_posix(). Since Subjobs inherit most of the parent job’s config we only output the input files and arguments that are specific to this subjob and no other details.
-
property
name¶ Getter for name of SubJob. Accesses the parent Job name to infer this.
-
property
output_dir¶ Getter for output_dir of SubJob. Accesses the parent Job output_dir to infer this.
-
parent= None¶ Job() instance of parent to this SubJob
-
ready()¶ Returns whether or not the Job has finished. If the job has subjobs then it will return true when they are all finished. It will return False as soon as it hits the first failure. Meaning that you cannot guarantee that all subjobs will have their status updated when calling this method. Instead use
update_status()to update all statuses if necessary.
-
result= None¶ The result object of this SubJob. Only filled once it is is submitted to a backend since the backend creates a special result class depending on its type.
-
property
status¶ Returns the status of this SubJob.
-
property
subjobs¶ A subjob cannot have subjobs. Always return empty list.
-
update_status()¶ Calls
update_status()on the job’s result. The result object should update all of the subjobs (if there are any) in the best way for the type of result object/backend.
-
property
working_dir¶ Getter for working_dir of SubJob. Accesses the parent Job working_dir to infer this.
-
SubJobSplitters and Arguments Generation¶
-
class
caf.backends.SubjobSplitter(*, arguments_generator=None)[source]¶ Abstract base class. This class handles the logic of creating subjobs for a
Jobobject. Thecreate_subjobsfunction should be implemented and used to construct the subjobs of the parent job object.- Parameters
arguments_generator (ArgumentsGenerator) – Used to construct the generator function that will yield the argument tuple for each
SubJob. The splitter will iterate through the generator each timecreate_subjobsis called. TheSubJobwill be sent into the generator withsend(subjob)so that the generator can decide what arguments to return.
-
arguments_generator= None¶ The
ArgumentsGeneratorused when creating subjobs.
-
assign_arguments(job)[source]¶ Use the
arguments_generator(if one exists) to assign the argument tuples to the subjobs.
-
class
caf.backends.MaxFilesSplitter(*, arguments_generator=None, max_files_per_subjob=1)[source]¶ -
assign_arguments(job)¶ Use the
arguments_generator(if one exists) to assign the argument tuples to the subjobs.
-
create_subjobs(job)[source]¶ This function creates subjobs for the parent job passed in. It creates as many subjobs as required in order to prevent the number of input files per subjob going over the limit set by
MaxFilesSplitter.max_files_per_subjob.
-
-
class
caf.backends.MaxSubjobsSplitter(*, arguments_generator=None, max_subjobs=1000)[source]¶ -
assign_arguments(job)¶ Use the
arguments_generator(if one exists) to assign the argument tuples to the subjobs.
-
create_subjobs(job)[source]¶ This function creates subjobs for the parent job passed in. It creates as many subjobs as required by the number of input files up to the maximum set by
MaxSubjobsSplitter.max_subjobs. If there are more input files thanmax_subjobsit instead groups files by the minimum number per subjob in order to respect the subjob limit e.g. If you have 11 input files and a maximum number of subjobs of 4, then it will create 4 subjobs, 3 of them with 3 input files, and one with 2 input files.
-
-
class
caf.backends.ArgumentsSplitter(*, arguments_generator=None, max_subjobs=None)[source]¶ Creates SubJobs based on the given argument generator. The generator will be called until a
StopIterationis issued. Be VERY careful to not accidentally give an infinite generator! Otherwise it will simply create SubJobs until you run out of memory. You can set theArgumentsSplitter.max_subjobsparameter to try and prevent this and throw an exception.This splitter is useful for MC production jobs where you don’t have any input files, but you want to control the exp/run numbers of subjobs. If you do have input files set for the parent
Jobobjects, then the same input files will be assinged to everySubJob.- Parameters
arguments_generator (ArgumentsGenerator) – The standard ArgumentsGenerator that is used to assign arguments
-
assign_arguments(job)¶ Use the
arguments_generator(if one exists) to assign the argument tuples to the subjobs.
-
create_subjobs(job)[source]¶ This function creates subjobs for the parent job passed in. It creates subjobs until the
SubjobSplitter.arguments_generatorfinishes.If
ArgumentsSplitter.max_subjobsis set, then it will throw an exception if more than this number of subjobs are created.
-
max_subjobs= None¶ If we try to create more than this many subjobs we throw an exception, if None then there is no maximum.
-
class
caf.backends.ArgumentsGenerator(generator_function, *args, **kwargs)[source]¶ -
args= None¶ Positional argument tuple used to ‘prime’ the
ArgumentsGenerator.generator_function.
-
property
generator¶ Returns: generator: The initialised generator (using the args and kwargs for initialisation). It should be ready to have
next/sendcalled on it.
-
generator_function= None¶ Generator function that has not been ‘primed’.
-
kwargs= None¶ Keyword argument dictionary used to ‘prime’ the
ArgumentsGenerator.generator_function.
-
Utility Functions + Objects¶
This module contains various utility functions for the CAF and Job submission Backends to use.
-
class
caf.utils.AlgResult[source]¶ Enum of Calibration results. Shouldn’t be very necessary to use this over the direct CalibrationAlgorithm members but it’s nice to have something pythonic ready to go.
-
failure= 3¶ failure Return code
-
iterate= 1¶ iteration required Return code
-
not_enough_data= 2¶ not enought data Return code
-
ok= 0¶ OK Return code
-
-
caf.utils.B2INFO_MULTILINE(lines)[source]¶ -
Quick little function that creates a string for B2INFO from a list of strings. But it appends a newline character + the necessary indentation to the follwing line so that the B2INFO output is nicely aligned. Then it calls B2INFO on the output.
-
class
caf.utils.CentralDatabase(global_tag)[source]¶ Simple class to hold the information about a bas2 Central database. Does no checking that a global tag exists. This class could be made much simpler, but it’s made to be similar to LocalDatabase.
- Parameters
global_tag (str) – The Global Tag of the central database
-
class
caf.utils.ExpRun[source]¶ Class to define a single (Exp,Run) number i.e. not an IoV. It is derived from a namedtuple created class.
We use the name ‘ExpRun_Factory’ in the factory creation so that the MRO doesn’t contain two of the same class names which is probably fine but feels wrong.
-
class
caf.utils.IoV(exp_low=- 1, run_low=- 1, exp_high=- 1, run_high=- 1)[source]¶ Python class to more easily manipulate an IoV and compare against others. Uses the C++ framework IntervalOfValidity internally to do various comparisons. It is derived from a namedtuple created class.
We use the name ‘IoV_Factory’ in the factory creation so that the MRO doesn’t contain two of the same class names which is probably fine but feels wrong.
Default construction is an ‘empty’ IoV of -1,-1,-1,-1 e.g. i = IoV() => IoV(exp_low=-1, run_low=-1, exp_high=-1, run_high=-1)
For an IoV that encompasses all experiments and runs use 0,0,-1,-1.
-
class
caf.utils.IoV_Result(iov, result)¶ -
property
iov¶ Alias for field number 0
-
property
result¶ Alias for field number 1
-
property
-
class
caf.utils.LocalDatabase(filepath, payload_dir='')[source]¶ Simple class to hold the information about a basf2 Local database. Does a bit of checking that the file path entered is valid etc.
- Paramters:
filepath (str): The file path of the database.txt file of the localdb
- Keyword Arguments
payload_dir (str) – If the payload directory is different to the directory containing the filepath, you can set it here.
-
class
caf.utils.PathExtras(path=None)[source]¶ Simple wrapper for basf2 paths to allow some extra python functionality directly on them e.g. comparing whether or not a module is contained within a path with ‘in’ keyword.
-
index(module_name)[source]¶ Returns the index of the first instance of a module in the contained path.
-
path= None¶ Attribute to hold path object that this class wraps
-
-
caf.utils.all_dependencies(dependencies, order=None)[source]¶ Here we pass in a dictionary of the form that is used in topological sort where the keys are nodes, and the values are a list of the nodes that depend on it.
However, the value (list) does not necessarily contain all of the future nodes that depend on each one, only those that are directly adjacent in the graph. So there are implicit dependencies not shown in the list.
This function calculates the implicit future nodes and returns an OrderedDict with a full list for each node. This may be expensive in memory for complex graphs so be careful.
If you care about the ordering of the final OrderedDict you can pass in a list of the nodes. The final OrderedDict then has the same order as the order parameter.
-
caf.utils.b2info_newline= '\n '¶ A newline string for B2INFO that aligns with the indentation of B2INFO’s first line
-
caf.utils.create_directories(path, overwrite=True)[source]¶ Creates a new directory path. If it already exists it will either leave it as is (including any contents), or delete it and re-create it fresh. It will only delete the end point, not any intermediate directories created.
-
caf.utils.decode_json_string(object_string)[source]¶ Simple function to call json.loads() on a string to return the Python object constructed (Saves importing json everywhere).
-
caf.utils.find_absolute_file_paths(file_path_patterns)[source]¶ Takes a file path list (including wildcards) and performs glob.glob() to extract the absolute file paths to all matching files.
Also uses set() to prevent multiple instances of the same file path but returns a list of file paths.
Any non “file” type urls are taken as absolute file paths already and are simply passed through.
-
caf.utils.find_gaps_in_iov_list(iov_list)[source]¶ Finds the runs that aren’t covered by the input IoVs in the list. This cannot find missing runs which lie between two IoVs that are separated by an experiment e.g. between IoV(1,1,1,10) => IoV(2,1,2,5) it is unknown if there were supposed to be more runs than run number 10 in experiment 1 before starting experiment 2. Therefore this is not counted as a gap and will not be added to the output list of IoVs
-
caf.utils.find_int_dirs(dir_path)[source]¶ If you previously ran a Calibration and are now re-running after failure, you may have iteration directories from iterations above your current one. This function will find directories that match an integer.
- Parameters
dir_path (
pathlib.Path) – The dircetory to search inside.- Returns
The matching Path objects to the directories that are valid ints
- Return type
list[
pathlib.Path]
-
caf.utils.find_run_lists_from_boundaries(boundaries, runs)[source]¶ Takes a list of starting ExpRun boundaries and a list of available ExpRuns and finds the runs that are contained in the IoV of each boundary interval. We assume that this is occuring in only one Experiment! We also assume that after the last boundary start you want to include all runs that are higher than this starting ExpRun. Note that the output ExpRuns in their lists will be sorted. So the ordering may be different than the overall input order.
-
caf.utils.find_sources(dependencies)[source]¶ Returns a deque of node names that have no input dependencies.
-
caf.utils.get_file_iov_tuple(file_path)[source]¶ Simple little function to return both the input file path and the relevant IoV, instead of just the IoV.
-
caf.utils.get_iov_from_file(file_path)[source]¶ Returns an IoV of the exp/run contained within the given file. Uses the b2file-metadata-show basf2 tool.
-
caf.utils.iov_from_runs(runs)[source]¶ Takes a list of (Exp,Run) and returns the overall IoV from the lowest ExpRun to the highest. It returns an IoV() object and assumes that the list was in order to begin with.
-
caf.utils.iov_from_runvector(iov_vector)[source]¶ Takes a vector of ExpRun from CalibrationAlgorithm and returns the overall IoV from the lowest ExpRun to the highest. It returns an IoV() object. It assumes that the vector was in order to begin with.
-
caf.utils.make_file_to_iov_dictionary(file_path_patterns, polling_time=10, pool=None, filterfalse=None)[source]¶ Takes a list of file path patterns (things that glob would understand) and runs b2file-metadata-show over them to extract the IoV.
- Paramters:
file_path_patterns (list[str]): The list of file path patterns you want to get IoVs for.
- Keyword Arguments
polling_time (int) – Time between checking if our results are ready.
pool – Optional Pool object used to multprocess the b2file-metadata-show subprocesses. We don’t close or join the Pool as you might want to use it yourself, we just wait until the results are ready.
filterfalse (function) – An optional function object that will be called on each absolute filepath found from your patterns. If True is returned the file will have its metadata returned. If False it will be skipped. The filter function should take the filepath string as its only argument.
- Returns
Mapping of matching input file paths (Key) to their IoV (Value)
- Return type
-
caf.utils.merge_local_databases(list_database_dirs, output_database_dir)[source]¶ Takes a list of database directories and merges them into one new directory, defined by the output_database_dir. It assumes that each of the database directories is of the standard form:
- directory_name
-> database.txt -> <payload file name> -> <payload file name> -> …
-
caf.utils.method_dispatch(func)[source]¶ Decorator that behaves exactly like functools.singledispatch but which takes the second argument to be the important one that we want to check the type of and dispatch to the correct function.
This is needed when trying to dispatch a method in a class, since the first argument of the method is always ‘self’. Just decorate around class methods and their alternate functions:
>>> @method_dispatch # Default method >>> def my_method(self, default_type, ...): >>> pass
>>> @my_method.register(list) # Registers list method for dispatch >>> def _(self, list_type, ...): >>> pass
Doesn’t work the same for property decorated class methods, as these return a property builtin not a function and change the method naming. Do this type of decoration to get them to work:
>>> @property >>> def my_property(self): >>> return self._my_property
>>> @my_property.setter >>> @method_dispatch >>> def my_property(self, input_property): >>> pass
>>> @my_property.fset.register(list) >>> def _(self, input_list_properties): >>> pass
-
caf.utils.pairwise(iterable)[source]¶ Iterate through a sequence by pairing up the current and next entry. Note that when you hit the last one you don’t get a (last, null), the final iteration gives you (last-1, last) and then finishes. If you only have one entry in the sequence this may be important as you will not get any looping.
- Parameters
iterable (list) – The iterable object we will loop over
- Returns
list[tuple]
-
caf.utils.parse_file_uri(file_uri)[source]¶ A central function for parsing file URI strings. Just so we only have to change it in one place later.
- Parameters
file_uri (str) –
- Returns
urllib.parse.ParseResult
-
caf.utils.parse_raw_data_iov(file_path)[source]¶ For as long as the Raw data is stored using a predictable directory/filename structure we can take advantage of it to more quickly infer the IoV of the files.
-
caf.utils.runs_from_vector(exprun_vector)[source]¶ Takes a vector of
ExpRunfrom CalibrationAlgorithm and returns a Python list of (exp,run) tuples in the same order.- Parameters
exprun_vector (
ROOT.vector[ROOT.pair(int,int)]) – Vector of expruns for conversion- Returns
list[ExpRun]
-
caf.utils.runs_overlapping_iov(iov, runs)[source]¶ Takes an overall IoV() object and a list of ExpRun and returns the set of ExpRun containing only those runs that overlap with the IoV.
-
caf.utils.temporary_workdir(path)[source]¶ Context manager that changes the working directory to the given path and then changes it back to its previous value on exit.
-
caf.utils.topological_sort(dependencies)[source]¶ Does a topological sort of a graph (dictionary) where the keys are the node names, and the values are lists of node names that depend on the key (including zero dependencies). It should return the sorted list of nodes.
>>> dependencies = {} >>> dependencies['c'] = ['a','b'] >>> dependencies['b'] = ['a'] >>> dependencies['a'] = [] >>> sorted = topological_sort(dependencies) >>> print(sorted) ['c', 'b', 'a']
Advanced Usage¶
There are several options for the CAF that most users will never need. But for more adventurous calibration developers there are quite a lot of configuration options and possibilities for extending the functionality of the CAF.
Writing Your Own Calibration Class¶
-
class
caf.framework.CalibrationBase(name, input_files=None)[source]¶ Bases:
abc.ABC,threading.ThreadAbstract base class of Calibration types. The CAF implements the
Calibrationclass which inherits from this and runs the C++ CalibrationCollectorModule and CalibrationAlgorithm classes. But by inheriting from this class and providing the minimal necessary methods/attributes you could plug in your own Calibration types that doesn’t depend on the C++ CAF at all and run everything in your own way.Warning
Writing your own class inheriting from
CalibrationBaseclass is not recommended! But it’s there if you really need it.- Parameters
name (str) – Name of this calibration object. Should be unique if you are going to run it.
- Keyword Arguments
input_files (list[str]) – Input files for this calibration. May contain wildcard expressions useable by
glob.glob.
-
property
daemon¶ A boolean value indicating whether this thread is a daemon thread.
This must be set before start() is called, otherwise RuntimeError is raised. Its initial value is inherited from the creating thread; the main thread is not a daemon thread and therefore all threads created in the main thread default to daemon = False.
The entire Python program exits when no alive non-daemon threads are left.
-
dependencies= None¶ List of calibration objects, where each one is a dependency of this one.
-
dependencies_met()[source]¶ Checks if all of the Calibrations that this one depends on have reached a successful end state.
-
depends_on(calibration)[source]¶ - Parameters
calibration (
CalibrationBase) – The Calibration object which will produce constants that this one depends on.
Adds dependency of this calibration on another i.e. This calibration will not run until the dependency has completed, and the constants produced will be used via the database chain.
You can define multiple dependencies for a single calibration simply by calling this multiple times. Be careful when adding the calibration into the
CAFnot to add a circular/cyclic dependency. If you do the sort will return an empty order and theCAFprocessing will fail.This function appens to the
CalibrationBase.dependenciesandCalibrationBase.future_dependenciesattributes of thisCalibrationBaseand the input one respectively. This prevents us having to do too much recalculation later on.
-
end_state= 'completed'¶ The name of the successful completion state. The
CAFwill use this as the state to decide when the Calibration is completed.
-
fail_state= 'failed'¶ The name of the failure state. The
CAFwill use this as the state to decide when the Calibration failed.
-
failed_dependencies()[source]¶ Returns the list of calibrations in our dependency list that have failed.
-
files_to_iovs= None¶ File -> Iov dictionary, should be
>>> {absolute_file_path:iov}
Where iov is a
IoVobject. Will be filled duringCAF.run()if empty. To improve performance you can fill this yourself before callingCAF.run()
-
future_dependencies= None¶ List of calibration objects that depend on this one.
-
property
ident¶ Thread identifier of this thread or None if it has not been started.
This is a nonzero integer. See the get_ident() function. Thread identifiers may be recycled when a thread exits and another thread is created. The identifier is available even after the thread has exited.
-
input_files= None¶ Files used for collection procedure
-
isAlive()¶ Return whether the thread is alive.
This method returns True just before the run() method starts until just after the run() method terminates. The module function enumerate() returns a list of all alive threads.
-
is_alive()¶ Return whether the thread is alive.
This method returns True just before the run() method starts until just after the run() method terminates. The module function enumerate() returns a list of all alive threads.
-
abstract
is_valid()[source]¶ A simple method you should implement that will return True or False depending on whether the Calibration has been set up correctly and can be run safely.
-
jobs_to_submit= None¶ A simple list of jobs that this Calibration wants submitted at some point.
-
join(timeout=None)¶ Wait until the thread terminates.
This blocks the calling thread until the thread whose join() method is called terminates – either normally or through an unhandled exception or until the optional timeout occurs.
When the timeout argument is present and not None, it should be a floating point number specifying a timeout for the operation in seconds (or fractions thereof). As join() always returns None, you must call isAlive() after join() to decide whether a timeout happened – if the thread is still alive, the join() call timed out.
When the timeout argument is not present or None, the operation will block until the thread terminates.
A thread can be join()ed many times.
join() raises a RuntimeError if an attempt is made to join the current thread as that would cause a deadlock. It is also an error to join() a thread before it has been started and attempts to do so raises the same exception.
-
output_database_dir= None¶ The directory where we’ll store the local database payloads from this calibration
-
abstract
run()[source]¶ The most important method. Runs inside a new Thread and is called from
CalibrationBase.startonce the dependencies of thisCalibrationBasehave returned with state == end_state i.e. “completed”.
-
save_payloads= None¶ Marks this Calibration as one which has payloads that should be copied and uploaded. Defaults to True, and should only be False if this is an intermediate Calibration who’s payloads are never needed.
-
start()¶ Start the thread’s activity.
It must be called at most once per thread object. It arranges for the object’s run() method to be invoked in a separate thread of control.
This method will raise a RuntimeError if called more than once on the same thread object.
Strategy Objects¶
-
class
caf.strategies.AlgorithmStrategy(algorithm)[source]¶ Base class for Algorithm strategies. These do the actual execution of a single algorithm on collected data. Each strategy may be quite different in terms of how fast it may be, how database payloads are passed between executions, and whether or not final payloads have an IoV that is independent to the actual runs used to calculates them.
- Parameters
algorithm (
caf.framework.Algorithm) – The algorithm we will run
This base class defines the basic attributes and methods that will be automatically used by the selected AlgorithmRunner. When defining a derived class you are free to use these attributes or to implement as much functionality as you want.
If you define your derived class with an __init__ method, then you should first call the base class
AlgorithmStrategy.__init__()method via super() e.g.>>> def __init__(self): >>> super().__init__()
The most important method to implement is
AlgorithmStrategy.run()which will take an algorithm and execute it in the required way defined by the options you have selected/attributes set.-
COMPLETED= 'COMPLETED'¶ Completed state
-
FAILED= 'FAILED'¶ Failed state
-
FINISHED_RESULTS= 'DONE'¶ Signal value that is put into the Queue when there are no more results left
-
algorithm= None¶ Algorithm() class that we’re running
-
allowed_granularities= ['run', 'all']¶ Granularity of collector that can be run by this algorithm properly
-
any_failed_iov()[source]¶ - Returns
If any result in the current results list has a failed algorithm code we return True
- Return type
-
database_chain= None¶ User defined database chain i.e. the default global tag, or if you have localdb’s/tags for custom alignment etc
-
dependent_databases= None¶ CAF created local databases from previous calibrations that this calibration/algorithm depends on
-
find_iov_gaps()[source]¶ Finds and prints the current gaps between the IoVs of the strategy results. Basically these are the IoVs not covered by any payload. It CANNOT find gaps if they exist across an experiment boundary. Only gaps within the same experiment are found.
- Returns
iov_gaps(list[IoV])
-
ignored_runs= None¶ Runs that will not be included in ANY execution of the algorithm. Usually set by Calibration.ignored_runs. The different strategies may handle the resulting run gaps differently.
-
input_files= None¶ Collector output files, will contain all files retured by the output patterns
-
is_valid()[source]¶ - Returns
Whether or not this strategy has been set up correctly with all its necessary attributes.
- Return type
-
output_database_dir= None¶ The output database directory for the localdb that the algorithm will commit to
-
output_dir= None¶ The algorithm output directory which is mostly used to store the stdout file
-
queue= None¶ The multiprocessing Queue we use to pass back results one at a time
-
required_attrs= ['algorithm', 'database_chain', 'dependent_databases', 'output_dir', 'output_database_dir', 'input_files', 'ignored_runs']¶ Required attributes that must exist before the strategy can run properly. Some are allowed be values that return False whe tested e.g. “” or []
-
required_true_attrs= ['algorithm', 'output_dir', 'output_database_dir', 'input_files']¶ Attributes that must have a value that returns True when tested by
is_valid().
-
results= None¶ The list of results objects which will be sent out before the end
SingleIoV¶
-
class
caf.strategies.SingleIOV(algorithm)[source]¶ Bases:
caf.strategies.AlgorithmStrategyThe fastest and simplest Algorithm strategy. Runs the algorithm only once over all of the input data or only the data corresponding to the requested IoV. The payload IoV is the set to the same as the one that was executed.
This uses a
caf.state_machines.AlgorithmMachineto actually execute the various steps rather than operating on a CalibrationAlgorithm C++ class directly.-
machine= None¶ caf.state_machines.AlgorithmMachineused to help set up and execute CalibrationAlgorithm It gets setup properly inrun()
-
run(iov, iteration, queue)[source]¶ Runs the algorithm machine over the collected data and fills the results.
-
usable_params= {'apply_iov': <class 'caf.utils.IoV'>}¶ The params that you could set on the Algorithm object which this Strategy would use. Just here for documentation reasons.
-
SequentialRunByRun¶
Fig. 9.3 Approximate execution logic of the SequentialRunByRun strategy¶
-
class
caf.strategies.SequentialRunByRun(algorithm)[source]¶ Bases:
caf.strategies.AlgorithmStrategyAlgorithm strategy to do run-by-run calibration of collected data. Runs the algorithm over the input data contained within the requested IoV, starting with the first run’s data only. If the algorithm returns ‘not enough data’ on the current run set, it won’t commit the payloads, but instead adds the next run’s data and tries again.
Once an execution on a set of runs return ‘iterate’ or ‘ok’ we move onto the next runs (if any are left) and start the same procedure again. Committing of payloads to the outputdb only happens once we’re sure that there is enough data in the remaining runs to get a full execution. If there isn’t enough data remaining, the last runs are merged with the previous successful execution’s runs and a final execution is performed on all remaining runs.
Additionally this strategy will automatically make sure that IoV gaps in your input data are covered by a payload. This means that there shouldn’t be any IoVs that don’t get a new payload by the end of runnning an iteration.
This uses a
caf.state_machines.AlgorithmMachineto actually execute the various steps rather than operating on a CalibrationAlgorithm C++ class directly.-
allowed_granularities= ['run']¶ Granularity of collector that can be run by this algorithm properly
-
apply_experiment_settings(algorithm, experiment)[source]¶ Apply experiment-dependent settings. This is the default version, which does not do anything. If necessary, it should be reimplemented by derived classes.
-
machine= None¶ caf.state_machines.AlgorithmMachineused to help set up and execute CalibrationAlgorithm It gets setup properly inrun()
-
run(iov, iteration, queue)[source]¶ Runs the algorithm machine over the collected data and fills the results.
-
usable_params= {'has_experiment_settings': <class 'bool'>, 'iov_coverage': <class 'caf.utils.IoV'>, 'step_size': <class 'int'>}¶ The params that you could set on the Algorithm object which this Strategy would use. Just here for documentation reasons.
-
SimpleRunByRun¶
-
class
caf.strategies.SimpleRunByRun(algorithm)[source]¶ Bases:
caf.strategies.AlgorithmStrategyAlgorithm strategy to do run-by-run calibration of collected data. Runs the algorithm over the input data contained within the requested IoV, starting with the first run’s data only.
This strategy differs from
SequentialRunByRunin that it will not merge run data if the algorithm returns ‘not enough data’ on the current run.Once an execution on a run returns any result ‘iterate’, ‘ok’, ‘not_enough_data’, or ‘failure’, we move onto the next run (if any are left). Committing of payloads to the outputdb only happens for ‘iterate’ or ‘ok’ return codes.
Important
Unlike most other strategies, this one won’t immediately fail and return if a run returns a ‘failure’ exit code. The failure will prevent iteration/successful completion of the CAF though.
Warning
Since this strategy doesn’t try to merge data from runs, if any run in your input data doesn’t contain enough data to complete the algorithm successfully, you won’t be able to get a successful calibration. The CAF then won’t allow you to iterate this calibration, or pass the constants onward to another calibration. However, you will still have the database created that covers all the successfull runs.
This uses a
caf.state_machines.AlgorithmMachineto actually execute the various steps rather than operating on a CalibrationAlgorithm C++ class directly.-
machine= None¶ caf.state_machines.AlgorithmMachineused to help set up and execute CalibrationAlgorithm It gets setup properly inrun()
-
run(iov, iteration, queue)[source]¶ Runs the algorithm machine over the collected data and fills the results.
-
usable_params= {}¶ The params that you could set on the Algorithm object which this Strategy would use. Just here for documentation reasons.
-
SequentialBoundaries¶
Fig. 9.4 Approximate execution logic of the SequentialBoundaries strategy¶
-
class
caf.strategies.SequentialBoundaries(algorithm)[source]¶ Bases:
caf.strategies.AlgorithmStrategyAlgorithm strategy to first calculate run boundaries where execution should be attempted. Runs the algorithm over the input data contained within the requested IoV of the boundaries, starting with the first boundary data only. If the algorithm returns ‘not enough data’ on the current boundary IoV, it won’t commit the payloads, but instead adds the next boundarie’s data and tries again. Basically the same logic as
SequentialRunByRunbut using run boundaries instead of runs directly. Notice that boundaries cannot span multiple experiments.By default the algorithm will get the payload boundaries directly from the algorithm that need to have inplemented the function
isBoundaryRequired. If the desired boundaries are already known it is possible to pass them directly setting the algorithm parameterpayload_boundariesand avoid the need to define theisBoundaryRequiredfunction.payload_boundariesis a list[(exp1, run1), (exp2, run2), ...]. A boundary at the beginning of each experiment will be added if not already present. An empty list will thus produce a single payload for each experiment. Apayload_boundariesset toNoneis equivalent to not passing it and restores the default behaviour where the boundaries are computed in theisBoundaryRequiredfunction of the algorithm.-
allowed_granularities= ['run']¶ Granularity of collector that can be run by this algorithm properly
-
execute_over_boundaries(boundary_iovs_to_run_lists, lowest_exprun, highest_exprun, iteration)[source]¶ Take the previously found boundaries and the run lists they correspond to and actually perform the Algorithm execution. This is assumed to be for a single experiment.
-
machine= None¶ caf.state_machines.AlgorithmMachineused to help set up and execute CalibrationAlgorithm It gets setup properly inrun()
-
run(iov, iteration, queue)[source]¶ Runs the algorithm machine over the collected data and fills the results.
-
usable_params= {'iov_coverage': <class 'caf.utils.IoV'>, 'payload_boundaries': []}¶ The params that you could set on the Algorithm object which this Strategy would use. Just here for documentation reasons.
-
State Machine Framework¶
-
class
caf.state_machines.AlgorithmMachine(algorithm=None, initial_state='init')[source]¶ Bases:
caf.state_machines.MachineA state machine to handle the logic of running the algorithm on the overall runs contained in the data.
-
add_state(state, enter=None, exit=None)¶ Adds a single state to the list of possible ones. Should be a unique string or a State object with a unique name.
-
add_transition(trigger, source, dest, conditions=None, before=None, after=None)¶ Adds a single transition to the dictionary of possible ones. Trigger is the method name that begins the transtion between the source state and the destination state.
The condition is an optional function that returns True or False depending on the current state/input.
-
algorithm= None¶ Algorithm() object whose state we are modelling
-
database_chain= None¶ Assigned database chain to the overall Calibration object, or to the ‘default’ Collection. Database chains for manually created Collections have no effect here.
-
static
default_condition(**kwargs)¶ Method to always return True.
-
default_states= None¶ Default states for the AlgorithmMachine
-
dependent_databases= None¶ CAF created local databases from previous calibrations that this calibration/algorithm depends on
-
get_transition_dict(state, transition)¶ Returns the transition dictionary for a state and transition out of it.
-
get_transitions(source)¶ Returns allowed transitions from a given state.
-
property
initial_state¶ The initial state of the machine. Needs a special property to prevent trying to run on_enter callbacks when set.
-
input_files= None¶ Collector output files, will contain all files retured by the output patterns
-
is_valid()[source]¶ - Returns
Whether or not this machine has been set up correctly with all its necessary attributes.
- Return type
-
output_database_dir= None¶ The output database directory for the localdb that the algorithm will commit to
-
output_dir= None¶ The algorithm output directory which is mostly used to store the stdout file
-
required_attrs= ['algorithm', 'dependent_databases', 'database_chain', 'output_dir', 'output_database_dir', 'input_files']¶ Required attributes that must exist before the machine can run properly. Some are allowed be values that return False whe tested e.g. “” or []
-
required_true_attrs= ['algorithm', 'output_dir', 'output_database_dir', 'input_files']¶ Attributes that must have a value that returns True when tested.
-
result= None¶ IoV_Result object for a single execution, will be reset upon a new execution
-
save_graph(filename, graphname)¶ Does a simple dot file creation to visualise states and transiitons.
-
setup_from_dict(params)[source]¶ - Parameters
params (dict) – Dictionary containing values to be assigned to the machine’s attributes of the same name.
-
property
state¶ The current state of the machine. Actually a
propertydecorator. It will call the exit method of the current state and enter method of the new one. To get around the behaviour e.g. for setting initial states, either use theinitial_stateproperty or directly set the _state attribute itself (at your own risk!).
-
-
class
caf.state_machines.CalibrationMachine(calibration, iov_to_calibrate=None, initial_state='init', iteration=0)[source]¶ Bases:
caf.state_machines.MachineA state machine to handle
Calibrationobjects and the flow of processing for them.-
add_state(state, enter=None, exit=None)¶ Adds a single state to the list of possible ones. Should be a unique string or a State object with a unique name.
-
add_transition(trigger, source, dest, conditions=None, before=None, after=None)¶ Adds a single transition to the dictionary of possible ones. Trigger is the method name that begins the transtion between the source state and the destination state.
The condition is an optional function that returns True or False depending on the current state/input.
-
automatic_transition()[source]¶ Automatically try all transitions out of this state once. Tries fail last.
-
calibration= None¶ Calibration object whose state we are modelling
-
collector_backend= None¶ Backend used for this calibration machine collector
-
static
default_condition(**kwargs)¶ Method to always return True.
-
default_states= None¶ States that are defaults to the
CalibrationMachine(could override later)
-
dependencies_completed()[source]¶ Condition function to check that the dependencies of our calibration are in the ‘completed’ state. Technically only need to check explicit dependencies.
-
files_containing_iov(file_paths, files_to_iovs, iov)[source]¶ Lookup function that returns all files from the file_paths that overlap with this IoV.
-
get_transition_dict(state, transition)¶ Returns the transition dictionary for a state and transition out of it.
-
get_transitions(source)¶ Returns allowed transitions from a given state.
-
property
initial_state¶ The initial state of the machine. Needs a special property to prevent trying to run on_enter callbacks when set.
-
iov_to_calibrate= None¶ IoV to be executed, currently will loop over all runs in IoV
-
iteration= None¶ Which iteration step are we in
-
root_dir= None¶ root directory for this Calibration
-
save_graph(filename, graphname)¶ Does a simple dot file creation to visualise states and transiitons.
-
property
state¶ The current state of the machine. Actually a
propertydecorator. It will call the exit method of the current state and enter method of the new one. To get around the behaviour e.g. for setting initial states, either use theinitial_stateproperty or directly set the _state attribute itself (at your own risk!).
-
-
exception
caf.state_machines.ConditionError[source]¶ Bases:
caf.state_machines.MachineErrorException for when conditions fail during a transition.
-
with_traceback()¶ Exception.with_traceback(tb) – set self.__traceback__ to tb and return self.
-
-
class
caf.state_machines.Machine(states=None, initial_state='default_initial')[source]¶ Bases:
objectBase class for a final state machine wrapper. Implements the framwork that a more complex machine can inherit from.
The
transitionsattribute is a dictionary of trigger name keys, each value of which is another dictionary of ‘source’ states, ‘dest’ states, and ‘conditions’ methods. ‘conditions’ should be a list of callables or a single one. A transition is valid if it goes from an allowed state to an allowed state. Conditions are optional but must be a callable that returns True or False based on some state of the machine. They cannot have input arguments currently.Every condition/before/after callback function MUST take
**kwargsas the only argument (exceptselfif it’s a class method). This is because it’s basically impossible to determine which arguments to pass to which functions for a transition. Therefore this machine just enforces that every function should simply take**kwargsand use the dictionary of arguments (even if it doesn’t need any arguments).This also means that if you call a trigger with arguments e.g.
machine.walk(speed=5)you MUST use the keyword arguments rather than positional ones. Somachine.walk(5)will not work.-
add_state(state, enter=None, exit=None)[source]¶ Adds a single state to the list of possible ones. Should be a unique string or a State object with a unique name.
-
add_transition(trigger, source, dest, conditions=None, before=None, after=None)[source]¶ Adds a single transition to the dictionary of possible ones. Trigger is the method name that begins the transtion between the source state and the destination state.
The condition is an optional function that returns True or False depending on the current state/input.
-
get_transition_dict(state, transition)[source]¶ Returns the transition dictionary for a state and transition out of it.
-
property
initial_state¶ Pointless docstring since it’s a property
-
save_graph(filename, graphname)[source]¶ Does a simple dot file creation to visualise states and transiitons.
-
property
state¶ The current state of the machine. Actually a
propertydecorator. It will call the exit method of the current state and enter method of the new one. To get around the behaviour e.g. for setting initial states, either use theinitial_stateproperty or directly set the _state attribute itself (at your own risk!).
-
states= None¶ Valid states for this machine
-
transitions= None¶ Allowed transitions between states
-
-
exception
caf.state_machines.MachineError[source]¶ Bases:
ExceptionBase exception class for this module.
-
with_traceback()¶ Exception.with_traceback(tb) – set self.__traceback__ to tb and return self.
-
-
class
caf.state_machines.State(name, enter=None, exit=None)[source]¶ Bases:
objectBasic State object that can take enter and exit state methods and records the state of a machine.
You should assign the self.on_enter or self.on_exit attributes to callback functions or lists of them, if you need them.
-
name= None¶ Name of the State
-
property
on_enter¶ Callback list when entering state
-
property
on_exit¶ Callback list when exiting state
-
-
exception
caf.state_machines.TransitionError[source]¶ Bases:
caf.state_machines.MachineErrorException for when transitions fail.
-
with_traceback()¶ Exception.with_traceback(tb) – set self.__traceback__ to tb and return self.
-
Runner Objects¶
-
class
caf.runners.AlgorithmsRunner(name)[source]¶ Bases:
caf.runners.RunnerBase class for
AlgorithmsRunnerclasses. Defines the necessary information that will be provided to everyAlgorithmsRunnerused by theframework.CAFAn
AlgorithmsRunnerwill be given a list offramework.Algorithmobjects defined during the setup of aframework.Calibrationinstance. TheAlgorithmsRunnerdescribes how to run each of thestrategies.AlgorithmStrategyobjects. As an example, assume that a singleframework.Calibrationwas given and list of twoframework.Algorithminstances to run.In this example the chosen
AlgorithmsRunner.run()is simple and just loops over the list ofcaf.framework.Algorithmcalling each one’scaf.strategies.AlgorithmStrategy.run()methods in order. Thereby generating a localdb with the only communication between thestrategies.AlgorithmStrategyinstances coming from the database payloads being available from one algorithm to the next.But you could imagine a more complex situation. The
AlgorithmsRunnermight take the firstframework.Algorithmand call itsAlgorithmStrategy.runfor only the first (exp,run) in the collected data. Then it might not commit the payloads to a localdb but instead pass some calculated values to the next algorithm to run on the same IoV. Then it might go back and re-run the first AlgorithmStrategy with new information and commit payloads this time. Then move onto the next IoV.Hopefully you can see that while the default provided
AlgorithmsRunnerandAlgorithmStrategyclasses should be good for most situations, you have lot of freedom to define your own strategy if needed. By following the basic requirements for the interface to theframework.CAFyou can easily plugin a different special case, or mix and match a custom class with default CAF ones.The run(self) method should be defined for every derived
AlgorithmsRunner. It will be called once and only once for each iteration of (collector -> algorithm).Input files are automatically given via the
framework.Calibration.output_patternswhich constructs a list of all files in the collector output directories that match the output_patterns. If you have multiple types of output data it is your job to filter through the input files and assign them correctly.A list of local database paths are given to the
AlgorithmsRunnerbased on theframework.Calibrationdependencies and any overall database chain given to the Calibration before running. By default you can call the “setup_algorithm” transition of thecaf.state_machines.AlgorithmMachineto automatically set a database chain based on this list. But you have freedom to not call this at all inrun, or to implement a different method to deal with this.-
algorithms= None¶ The list of algorithms that this runner executes
-
database_chain= None¶ User set databases, can be used to apply your own constants and global tags
-
dependent_databases= None¶ List of local databases created by previous CAF calibrations/iterations
-
final_state= None¶ Final state of runner
-
input_files= None¶ All of the output files made by the collector job and recovered by the “output_patterns”
-
name= None¶ The name of this runner instance
-
output_database_dir= None¶ The directory of the local database we use to store algorithm payloads from this execution
-
output_dir= None¶ Output directory of these algorithms, for logging mostly
-
results= None¶ Algorithm results from each algorithm we execute
-
abstract
run()¶
-
-
exception
caf.runners.RunnerError[source]¶ Bases:
ExceptionBase exception class for Runners
-
with_traceback()¶ Exception.with_traceback(tb) – set self.__traceback__ to tb and return self.
-
-
class
caf.runners.SeqAlgorithmsRunner(name)[source]¶ Bases:
caf.runners.AlgorithmsRunner
Skimming Events With The SoftwareTriggerResult¶
Each event recorded by Belle II must have passed the High Level Trigger (HLT). The HLT runs some reconstruction online and applies cuts on the reconstructed objects in order to to make decisions about which events to keep. The decisions are made by the SoftwareTrigger Module and stored in the output basf2 file in a SoftwareTriggerResult object.
Certain calibration analyses require broad types of events (di-muon, Bhabha, …) which can be skimmed out of the basf2 file
by requiring that some HLT trigger decisions were True/False.
The TriggerSkim Module is provided to help extract the decisions stored in the SoftwareTriggerResult object.
You can then use the if_value() to decide what to do with the returned value of the
TriggerSkim Module.
-
TriggerSkim¶ Trigger Skim Module
Module will return an overall result (True/False) based on whether or not ANY or ALL (depending on
logicMode) of the specified triggers returned the given value (expectedResult) from the SoftwareTriggerResult.Optionally it can apply prescales on each trigger line. In that case
logicMode='and'might not be very meaningful as the return value will only be true if all trigger lines were accepted after their own prescale.- Package
calibration
- Library
libTriggerSkim.so
- Required Parameters
- triggerLines (list(variant(str, tuple(str, unsigned int))))
Trigger lines to skim on. This can either be just a list of names or each item can also optionally be a tuple with name and prescale factor. So either
['hadron', 'cosmic']or[('hadron', 1), ('cosmic', 20)]or a mix of both
- Parameters
- expectedResult (int, default=1)
The SoftwareTriggerResult value that each trigger line in the
triggerLinesparameter will be tested for.
- logicMode (str, default=’or’)
Each trigger line in
triggerLineswill be tested against theexpectedResultparameter. The logic mode controls whether we test using ‘and’ or ‘or’ logic to get a return value. ‘and’ means that all trigger lines must haveresults == expectedResult, ‘or’ means that only one of the trigger lines must match the expectedResult value.
- resultOnMissing (int or None, default=None)
Value to return if there’s no hlt trigger result available. If this is set to None a FATAL error will be raised if the results are missing. Otherwise the value given will be set as return value of the module
- useRandomNumbersForPreScale (bool, default=True)
Flag to use random numbers (True) or a counter (False) for applying the prescale. In the latter case, the module will retain exactly one event every N processed, where N (the counter value) is set for each line via the
triggerLinesoption. By default, random numbers are used.