First steering file
Contents
3.4.2. First steering file#
In this hands-on tutorial you’ll be writing your first steering file. Our ultimate goal is to reconstruct \(B^0 \to J/\Psi(\to e^+e^-)K_S^0(\to \pi^+\pi^+)\). You’ll be learning step-by-step what is necessary to achieve this, and in the end you will produce a plot of the \(B\) meson candidates. As you have already learned in the previous sections, basf2 provides a large variety of functionality. While the final steering file of this lesson will be working and producing some reasonable output, there are many possible extensions that you will learn all about in the succeeding lessons.
Let’s get started: The very first step is always to set up the necessary environment.
Task
Set up the basf2 environment using the currently recommended software version.
Hint
Go back to The basics. to see the step-by-step instructions.
Solution
source /cvmfs/belle.cern.ch/tools/b2setup
b2help-releases
b2setup <release-shown-at-prompt-by-previous-script>
Now let’s get started with your steering file!
Task
Open an empty file with an editor of your choice. Add three lines that do the following:
Import the
basf2python library (it might be convenient to set an abbreviation, e.g.b2)Create a
basf2.Path(call itmain)Process the path with
basf2.process
Save the file as myanalysis.py.
Hint
Have a look at basf2 and basf2.process
Solution
import basf2 as b2
main = b2.Path()
b2.process(main)
Running steering files is as easy as calling basf2 myanalysis.py on the
command-line.
Exercise
Run the short script that you just created. Don’t worry, if you’ve done everything correct, you should see some error messages. Read them carefully!
Solution
The output should look like this:
[INFO] Steering file: myanalysis.py
[INFO] Starting event processing, random seed is set to '94887e3828c78b3bd0b761678bd255317f110e183c2ed59ebdcd027e7610b9d6'
[ERROR] There is no module that provides event and run numbers (EventMetaData). You must add either the EventInfoSetter or an input module (e.g. RootInput) to the beginning of your path.
[FATAL] 1 ERROR(S) occurred! The processing of events will not be started. { function: void Belle2::EventProcessor::process(const PathPtr&, long int) }
[INFO] ===Error Summary================================================================
[FATAL] 1 ERROR(S) occurred! The processing of events will not be started.
[ERROR] There is no module that provides event and run numbers (EventMetaData). You must add either the EventInfoSetter or an input module (e.g. RootInput) to the beginning of your path.
[INFO] ================================================================================
[ERROR] in total, 1 errors occurred during processing
The random seed will of course differ in your case.
Loading input data#
Of course, no events could be processed so far because no data has been loaded
yet. Let’s do it. As already described in the previous lesson almost all
convenience functions that are needed can be found in modularAnalysis.
It is recommended to use inputMdstList or inputMdst. If you look at the
source code, you’ll notice that the latter actually calls the more general
former.
Exercise
How many arguments are required for the inputMdstList function?
Which value has to be set for the environment type?
Hint
Take a look at the signature of the function. It is highlighted in
blue at the top. Some arguments take a default value, e.g. skipNEvents=0
and are therefore not required.
Solution
Two parameters have no default value and are therefore required:
a list of root input files
the path
The environmentType only has to be modified if you are analyzing Belle
data / MC.
In a later lesson you’ll learn how and where to find input files for your
analysis. For the purpose of this tutorial we have prepared some local input
files of \(B^0 \to J/\Psi K_S^0\). They should be available in the
${BELLE2_EXAMPLES_DATA_DIR}/starterkit/2021 directory on KEKCC, NAF, and
other servers. The files’ names start with the decfile number 1111540100.
If you’re working from an institute server
Perhaps you are working on the server of your home institute and this folder
is not available. In this case please talk to your administrators to make
the ${BELLE2_EXAMPLES_DATA_DIR} available for you and make sure that it
is synchronized. Note that we might not be able to provide you with the
same level of support on other machines though.
If you’re working on your own machine
In this case you might first need to copy the data files to your home
directory on your local machine from kekcc or DESY via an SSH connection (cf.
SSH - Secure Shell) and then either change the path accordingly or set
the BELLE2_EXAMPLES_DATA_DIR environment variable to point to the right
directory. Note that we might not be able to provide you with the same level
of support on other machines though.
Exercise
Check out the location of the files mentioned above. Which two settings of MC are provided?
Hint
Remember the ls (and cd) bash command?
Solution
ls ${BELLE2_EXAMPLES_DATA_DIR}/starterkit/2021
Alternatively, you can first navigate to the directory with cd and then
just call ls without any arguments.
There are each 10 files with and without beam background (BGx1 and BGx0). Their names only differ by the final suffix, which is an integer between 0 and 9.
A helpful function to get common data files from the examples directory is
basf2.find_file.
Task
Extend your steering file by loading the data of one of the local input files. It makes sense to run the steering file again.
If there is a syntax error in your script or you forgot to include a necessary argument, there will be an error message that should help you to debug and figure out what needs to be fixed.
If the script is fine, only three lines with info messages should be printed to the output and you should see a quickly finishing progress bar.
Hint
Don’t forget to import modularAnalysis (this is the module that contains
inputMdstList). It might be convenient to set an abbreviation, e.g.
ma. Then you have to set the correct values for the two required
arguments of inputMdstList.
Solution
#!/usr/bin/env python3
import basf2 as b2
import modularAnalysis as ma
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file("starterkit/2021/1111540100_eph3_BGx0_0.root", "examples")],
path=main
)
# start the event loop (actually start processing things)
b2.process(main)
In the solution to the last task we have added empty lines, some comments, and used shortcuts for the imports. This helps to give the script a better structure and allows yourself and others to better understand what’s going on in the steering file. In the very first line we have also added a shebang to define that the steering file should be executed with a python interpreter.
So far, the input file has been completely hard-coded. But as we’ve seen before the file names only differ by the final suffix. We can be a little bit more flexibility by providing this integer as a command-line argument. Then, we can select a different input file when running the steering file, and without having to change anything in the script itself.
Task
Adjust your steering file so that you can select via an integer as a command-line argument which file is going to be processed.
Hint
You should have learned about command-line arguments in this
part of the python introduction from Software Carpentry. Otherwise, go
back and refresh your memory. All you have to do is to import the system
library, store the correct command-line argument (from sys.argv) in a local
variable filenumber, and extend the path string to include it.
Hint
You can get the integer from the command line arguments using
Hint
Rather than concatenating strings with + ("file_" + str(filenumber) + ".root"),
you can also use so-called f-strings: f"file_{filenumber}.root". They
are great for both readability and performance.
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# start the event loop (actually start processing things)
b2.process(main)
Tip
Make sure that from now on you always supply a number every time you run your
steering file, e.g. basf2 myanalysis.py 1.
Otherwise you will get an exception like this
Traceback (most recent call last):
File "myanalysis.py", line 3, in <module>
filenumber = sys.argv[1]
IndexError: list index out of range
Filling particle lists#
The mdst data objects (Tracks, ECLCluster, KLMCluster, V0s) of the input file
have to be transferred into Particle data objects. This is done via the
ParticleLoader module and its wrapper function (convenience function) fillParticleList.
Exercise
Read the documentation of fillParticleList to familiarize yourself with
the required arguments.
Which six final state particles can be created from Tracks?
Solution
Electrons, muons, pions, kaons, protons, and deuterons.
Internally, the anti-particle lists are always filled as well, so it is not
necessary to call fillParticleList for e+ and e-. In fact, you will
see a warning message for the second call telling you that the corresponding
particle list already exists.
As long as no selection criteria (cuts) are provided, the only difference between loading different charged final state particle types is the mass hypothesis used in the track fit.
Each particle used in the decayString argument of the fillParticleList
function can be extended with a label. This is useful to distinguish between
multiple lists of the same particle type with different selection criteria,
e.g. soft and hard electrons.
ma.fillParticleList("e-:soft", "E < 1", path=main) # the label of this electron list is "soft"
ma.fillParticleList("e-:hard", "E > 3", path=main) # here the label is "hard"
Warning
If the provided cut string is not empty you can not use the label
all, i.e. having
ma.fillParticleList("e-:all", "E > 0", path=main)
in your steering file will cause a fatal error and stop the execution of your script.
There are standard particle lists with predefined selection criteria. While
those for charged final state particles should only be used in the early stages of
your analysis and be replaced with dedicated selections adjusted to the needs
of the decay mode you are studying, it is recommended to use them for V0s
(\(K_S^0\), \(\Lambda^0\)). They are part of the library stdV0s.
Exercise
Find the documentation of the convenience function that creates the standard \(K_S^0\) particle list. What is the name of the particle list generated by this function?
Hint
The documentation is here: stdV0s.stdKshorts.
Solution
It’s K_S0:merged because it is a combination of \(K_S^0\) candidates created
directly from V0s found in the tracking and combinations of two charged
pions.
Task
Extend your steering file by loading electrons, positrons, and \(K_S^0\) candidates. At
the very end of your script you should also print a summary table of all the
modules added to your path using the function statistics.
Hint
All you need is fillParticleList, stdKshorts, and statistics.
Remember that charge-conjugated particles are automatically created.
You do not need a cut for the electrons, so you can use an empty string
"".
Hint
Always keep in mind from which module your functions are taken.
Since stdKshorts comes from the module stdV0s, you need to import
it first.
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
import stdV0s
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# fill final state particle lists
ma.fillParticleList("e+:uncorrected", "", path=main)
stdV0s.stdKshorts(path=main)
# Start the event loop (actually start processing things)
b2.process(main)
# print out the summary
print(b2.statistics)
In the solution we gave the electrons the label uncorrected. This is
already in anticipation of a future extension in which Bremsstrahlung
recovery will be applied (Various additions).
Task
Run your steering file and answer the following questions:
Which are the mass window boundaries set for the \(K_S^0\)?
Which module had the longest execution time?
Hint
Don’t forget to include a number, e.g. 1 as a command line argument to
specify the input file number!
Solution
basf2 myanalysis.py 1
In the output there are INFO messages that the \(K_S^0\) invariant mass has
to be between 0.45 and 0.55 GeV/c 2.
The module TreeFitter_K_S0:RD takes the longest. It’s a vertex fit of
the \(K_S^0\) candidates. You will learn more about vertex fits in
Vertex fitting.
In the previous task you should have learned how useful it is to carefully study the output. This is especially relevant if there are warning or error messages. Remember to never ignore them as they usually point to some serious issue, either in the way you have written your steering file or in the basf2 software itself. In the latter case you are encouraged to report the problem so that it can be fixed by some experts (maybe you yourself will become this expert one day).
In order to purify a sample it makes sense to apply at least loose selection
criteria. This can be based on the particle identification (e.g. electronID
for electrons and positrons), requiring the tracks to originate from close to the
interaction point (by using dr and dz), and having a polar angle in the acceptance
of the CDC (thetaInCDCAcceptance).
Exercise
Find out what’s the difference between dr and dz, e.g. why do we
not have to explicitly ask for the absolute value of dr? What’s the angular
range of the CDC acceptance (as implemented in the software)?
Hint
The documentation of dr and dz should tell you all about the first
question. The angular range is a bit trickier. You have to directly
inspect the source code of the variable defined in the variables folder of
the analysis package. There has been an exercise on how to find the source
code in Getting started, and getting help interactively.
Solution
The variable dr gives the transverse distance, while dz is the
z-component of the point of closest approach (POCA) with respect to the
interaction point (IP). Components are signed, while distances are
magnitudes.
The polar range of the CDC acceptance is \(17^\circ < \theta < 150^\circ\) as written here.
Task
Apply a cut on the electron particle list, requiring an electron ID greater than 0.1, a maximal transverse distance to the IP of 0.5 cm, a maximal distance in z-direction to the IP of 2 cm, and the track to be inside the CDC acceptance.
Hint
Previously we were using an empty string "" as argument to
fileParticleList. Now you need to change this.
Solution
ma.fillParticleList( # [S10]
"e+:uncorrected",
"electronID > 0.1 and dr < 0.5 and abs(dz) < 2 and thetaInCDCAcceptance",
path=main,
) # [E10]
Note
Marker comments in the solution code
If you are wondering about comments like [S10] or [E10] in the code
snippets that we include in our tutorials: You can completely ignore them.
In case you are curious about their meaning: Sometimes we want to show only
a small part of a larger file. Copying the code or referencing the lines by
line numbers is a bit unstable (what if someone changes the code and
forgets to update the rest of the documentation?), so we use these tags to
mark the beginning and end of a subset of lines to be included in the
documentation.
Combining particles#
Now we have a steering file in which final state particles are loaded from the
input mdst file to particle lists. One of the most powerful modules of the
analysis software is the ParticleCombiner. It takes those particle lists and
finds all unique combinations. The same particle can of course not be used
twice, e.g. the two positive pions in \(D^0 \to K^- \pi^+ \pi^+ \pi^-\)
have to be different mdst track objects. However, all of this is taken care of
internally. For multi-body decays like the one described above, there can
easily be many multiple candidates which share some particles but differ by
at least one final state particle.
The wrapper function for the ParticleCombiner is
called reconstructDecay. Its first argument is a DecayString, which is a
combination of a mother particle (list), an arrow, and daughter particles. The
DecayString has its own grammar with several markers, keywords, and arrow
types. It is especially useful for inclusive reconstructions (reconstructions
in which only part of the decay products are specified, e.g. only requiring
charged leptons in the final state; the opposite would be exclusive reconstructions). Follow the
provided link if you want to learn more about the DecayString. For the
purpose of this tutorial, we do not need any of those fancy extensions as the
default arrow type -> suffices. However, it is important to know how the
particles themselves need to be written in the decay string.
Exercise
How do we have to type a \(J/\Psi\), and what is its nominal mass?
Hint
Make use of the tool b2help-particles
(basf2 particles). As a spin-1
\(c\bar{c}\) resonance the PDG code of the \(J/\Psi\) is 443.
Solution
The \(J/\Psi\) has to be typed J/psi. Whenever you misspell a
particle name in a decay string, there will be an error message telling
you that it is unknown.
The invariant mass of the \(J/\Psi\) is set to 3.0969 GeV/c 2.
Task
Extend the steering file by first forming \(J/\Psi\) candidates from electron-positron combinations, and then combining them with a \(K_S^0\) to form \(B^0\) candidates.
Include a abs(dM) < 0.11 cut for the \(J/\Psi\).
Hint
All you need is to call reconstructDecay twice.
Hint
The \(J/\Psi\) reconstruction looks like this:
# combine final state particles to form composite particles [S20]
ma.reconstructDecay(
"J/psi:ee -> e+:uncorrected e-:uncorrected", cut="abs(dM) < 0.11", path=main
) # [E20]
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
import stdV0s
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# fill final state particle lists
ma.fillParticleList( # [S10]
"e+:uncorrected",
"electronID > 0.1 and dr < 0.5 and abs(dz) < 2 and thetaInCDCAcceptance",
path=main,
) # [E10]
stdV0s.stdKshorts(path=main)
# combine final state particles to form composite particles [S20]
ma.reconstructDecay(
"J/psi:ee -> e+:uncorrected e-:uncorrected", cut="abs(dM) < 0.11", path=main
) # [E20]
# combine J/psi and KS candidates to form B0 candidates
ma.reconstructDecay(
"B0 -> J/psi:ee K_S0:merged",
cut="",
path=main,
)
# [E30]
Writing out information to an ntuple#
To separate signal from background events, and to extract physics parameters, an offline
analysis has to be performed. The final step of the steering file is to write
out information in a so called ntuple using variablesToNtuple. It can
contain one entry per candidate or one entry per event.
Exercise
How do you switch between the two ntuple modes?
Hint
Look at the documentation of variablesToNtuple.
Solution
When providing an empty decay string, an event-wise ntuple will be created.
Warning
Only variables declared as Eventbased are allowed in the event mode.
Conversely, both candidate and event-based variables are allowed in the
candidate mode.
A good variable to start with is the beam-constrained mass Mbc, which is defined
as
For correctly reconstructed \(B\) mesons this variable should peak at the \(B\) meson mass.
Task
Add some code that saves the beam-constrained \(B\) mass of each \(B\) candidate in an output ntuple. Then, run your steering file.
Hint
The variable for the beam-constrained \(B\) mass is called Mbc. It has to be
provided as an element of a list to the argument variables of the
variablesToNtuple function.
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
import stdV0s
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# fill final state particle lists
ma.fillParticleList( # [S10]
"e+:uncorrected",
"electronID > 0.1 and dr < 0.5 and abs(dz) < 2 and thetaInCDCAcceptance",
path=main,
) # [E10]
stdV0s.stdKshorts(path=main)
# combine final state particles to form composite particles [S20]
ma.reconstructDecay(
"J/psi:ee -> e+:uncorrected e-:uncorrected", cut="abs(dM) < 0.11", path=main
) # [E20]
# combine J/psi and KS candidates to form B0 candidates
ma.reconstructDecay(
"B0 -> J/psi:ee K_S0:merged",
cut="",
path=main,
)
# [E30]
# save variables to an output file (ntuple)
ma.variablesToNtuple(
"B0",
variables=['Mbc'],
filename="Bd2JpsiKS.root",
treename="tree",
path=main,
)
# start the event loop (actually start processing things)
b2.process(main)
# print out the summary
print(b2.statistics)
Although you are analyzing a signal MC sample, the reconstruction will find many candidates that are actually not signal, but random combinations that happen to fulfill all your selection criteria.
Task
Write a short python script in which you load the root ntuple from the previous exercise into a dataframe and then plot the distribution of the beam-constrained mass using a histogram with 100 bins between the range 4.3 to 5.3 GeV/c 2.
Can you identify the signal and background components?
Hint
Read in the ntuple using uproot. Use the
histogram plotting routine of the dataframe.
For instructions on the basic usage of uproot see the Getting started guide.
Hint
You might take a look back at your python training. This is a good use case
for a jupyter notebook. Make sure to include the %matplotlib inline
magic to see the plots.
Solution
import matplotlib.pyplot as plt
import uproot
df = uproot.open('Bd2JpsiKS.root:tree').arrays(['Mbc'], library='pd')
df.hist('Mbc', bins=100, range=(4.3, 5.3))
plt.xlabel(r'M$_{\rm bc}$ [GeV/c$^{2}$]')
plt.ylabel('Number of candidates')
plt.xlim(4.3, 5.3)
plt.savefig('Mbc_all.png')
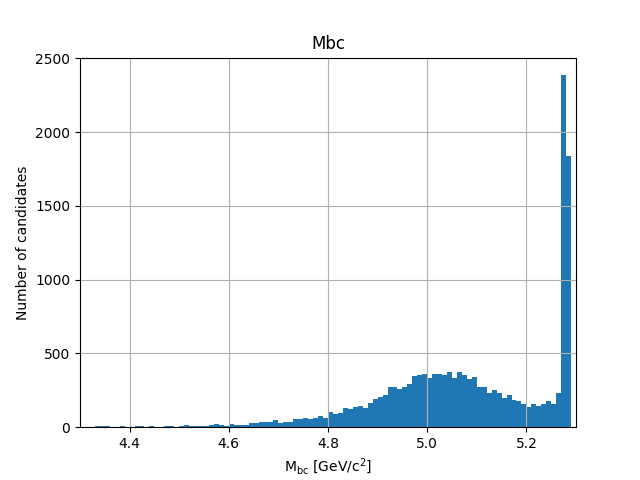
Fig. 3.21 There is a (signal) peak at the nominal \(B\) mass of 5.28 GeV/c 2 and lots of background candidates as a broad bump left of the peak.#
Adding MC information#
For the beam-constrained mass we know pretty well how the signal distribution should look like. But what’s the resolution and how much background actually extends under the signal peak? With MC, we have the advantage that we know what has been generated. Therefore, we can add a flag to every candidate to classify it as signal or background. Furthermore, we can study our background sources if we know what the reconstruction has falsely identified.
There is a long chapter on MC matching in the documentation. You should definitely read it to understand at least the basics.
Exercise
Which module do you have to run to get the relations between the reconstructed and the generated particles? How often do you have to call the corresponding function?
Hint
Did you have a look at the documentation of MC matching?
Solution
You need to run the MCMatcherParticles module, most conveniently
available via the wrapper function modularAnalysis.matchMCTruth. If this is
run on the head of the decay chain, it only needs to be called once because
the relations of all (grand)^N-daughter particles are set recursively.
Task
Add MC matching for all particles of the decay chain, and save information on whether the reconstructed \(B\) meson is a signal candidate to the ntuple. Run the steering file again.
Hint
Only one line of code is needed to call matchMCTruth.
Hint
Which variable is added by matchMCTruth? Remember to add it to the
ntuple!
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
import stdV0s
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# fill final state particle lists
ma.fillParticleList(
"e+:uncorrected",
"electronID > 0.1 and dr < 0.5 and abs(dz) < 2 and thetaInCDCAcceptance",
path=main,
)
stdV0s.stdKshorts(path=main)
# combine final state particles to form composite particles
ma.reconstructDecay(
"J/psi:ee -> e+:uncorrected e-:uncorrected", cut="abs(dM) < 0.11", path=main
)
# combine J/psi and KS candidates to form B0 candidates
ma.reconstructDecay(
"B0 -> J/psi:ee K_S0:merged",
cut="",
path=main,
)
# match reconstructed with MC particles
ma.matchMCTruth("B0", path=main)
# save variables to an output file (ntuple)
ma.variablesToNtuple(
"B0",
variables=['Mbc', 'isSignal'],
filename="Bd2JpsiKS.root",
treename="tree",
path=main,
)
# start the event loop (actually start processing things)
b2.process(main)
# print out the summary
print(b2.statistics)
Task
Plot the beam-constrained mass but this time use the signal flag to visualize which component is signal and which is background.
Hint
Remember the by keyword for df.hist? Alternatively you could also
use df.query. The necessary variable is called isSignal.
Solution
import matplotlib.pyplot as plt
import uproot
df = uproot.open('Bd2JpsiKS.root:tree').arrays(['isSignal', 'Mbc'], library='pd')
df.hist('Mbc', bins=100, range=(4.3, 5.3), by='isSignal')
plt.xlabel(r'M$_{\rm bc}$ [GeV/c$^{2}$]')
plt.ylabel('Number of candidates')
plt.xlim(4.3, 5.3)
plt.savefig('Mbc_MCsplit.png')
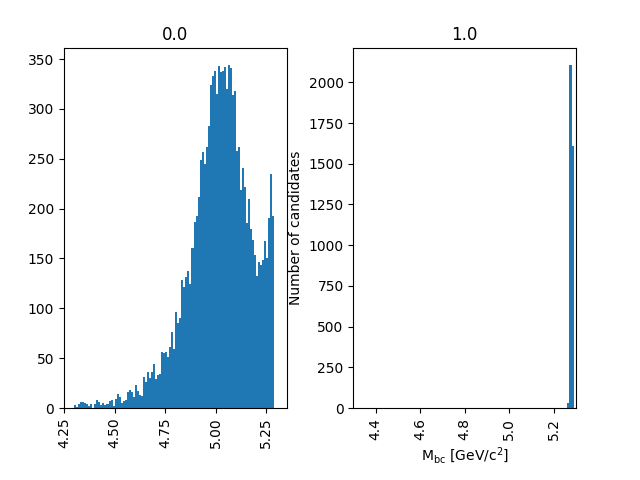
Fig. 3.22 The background peaks around 5 GeV/c 2, but does also extend into the signal peak region.#
As you could see, it makes sense to cut on Mbc from below. A complementary
variable that can be used to cut away background is \(\Delta E\) (deltaE).
Exercise
When combining your \(J/\Psi\) with your \(K_S^0\), introduce a cut \(\text{M}_{\rm bc} > 5.2\) and \(|\Delta E|<0.15\).
Hint
Take a look at the cut argument for reconstructDecay. You will also
need the abs metafunction.
Solution
# combine J/psi and KS candidates to form B0 candidates [S40]
ma.reconstructDecay(
"B0 -> J/psi:ee K_S0:merged",
cut="Mbc > 5.2 and abs(deltaE) < 0.15",
path=main,
) # [E40]
Variable collections#
While the MC matching allows us to separate signal from background and study their shapes, we need to use other variables to achieve the same on collision data. Initially, it makes sense to look at many different variables and try to find those with discriminating power between signal and background. The most basic information are the kinematic properties like the energy and the momentum (and its components). In basf2, collections of variables for several topics are pre-prepared. You can find the information in the Collections and Lists section of the documentation.
Exercise
Find out which variable collections contain the variables we have added to the ntuple so far.
Solution
The collection deltae_mbc contains Mbc and deltaE. The isSignal
variable along with many other truth match variables is in the collection
mc_truth.
Task
Save all the kinematic information, both the truth and the reconstructed values, of the \(B\) meson to the ntuple. Also, use the variable collections from the last exercise to replace the individual list.
Hint
The variable collections kinematics, mc_kinematics,
deltae_mbc, and mc_truth make your life a lot easier.
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
import stdV0s
import variables.collections as vc
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# fill final state particle lists
ma.fillParticleList(
"e+:uncorrected",
"electronID > 0.1 and dr < 0.5 and abs(dz) < 2 and thetaInCDCAcceptance",
path=main,
)
stdV0s.stdKshorts(path=main)
# combine final state particles to form composite particles
ma.reconstructDecay(
"J/psi:ee -> e+:uncorrected e-:uncorrected", cut="abs(dM) < 0.11", path=main
)
# combine J/psi and KS candidates to form B0 candidates [S40]
ma.reconstructDecay(
"B0 -> J/psi:ee K_S0:merged",
cut="Mbc > 5.2 and abs(deltaE) < 0.15",
path=main,
) # [E40]
# match reconstructed with MC particles
ma.matchMCTruth("B0", path=main)
# Create list of variables to save into the output file
b_vars = []
standard_vars = vc.kinematics + vc.mc_kinematics + vc.mc_truth
b_vars += vc.deltae_mbc
b_vars += standard_vars
# save variables to an output file (ntuple)
ma.variablesToNtuple(
"B0",
variables=b_vars,
filename="Bd2JpsiKS.root",
treename="tree",
path=main,
)
# start the event loop (actually start processing things)
b2.process(main)
# print out the summary
print(b2.statistics)
Hint
If you have trouble understanding what we are doing with the b_vars
list, simply add a couple of print(b_vars) between the definition and
the operations on it. You might also want to take another look at your
understanding of lists.
Variable aliases#
Apart from variables for the mother \(B\) meson, we are also interested in the
information of the other daughter and granddaughter variables. You can access
them via the daughter meta variable, which takes an integer and a variable
name as input arguments. The integer (0-based) counts through the daughter
particles, e.g. daughter(0, p) would be the momentum of the first
daughter, in our case of the \(J/\Psi\). This function can also be used
recursively.
Exercise
What does daughter(0, daughter(0, E)) denote?
Solution
It’s the energy of the positive electron.
In principle, one can add these nested variables directly to the ntuple,
but the brackets have to be escaped (i.e. replaced with “normal” characters),
and the resulting variable name in the ntuple is not very user-friendly or
intuitive. For example daughter(0, daughter(0, E)) becomes
daughter__bo0__cm__spdaughter__bo0__cm__spE__bc__bc. Not exactly pretty,
right?
So instead, let’s define aliases to translate the variable names!
This can be done with addAlias.
Exercise
How can you replace daughter(0, daughter(0, E)) with ep_E?
Hint
Check the documentation of addAlias for the correct syntax.
Solution
from variables import variables as vm
vm.addAlias("ep_E", "daughter(0, daughter(0, E))")
However, this can quickly fill up many, many lines. Therefore, there are utils
to easily create aliases. The most useful is probably
create_aliases_for_selected. It lets you select particles from a decay
string via the ^ operator for which you want to define aliases, and also
set a prefix. Another utility is create_aliases, which is particularly
useful to wrap a list of variables in another meta-variable like useCMSFrame
or matchedMC.
Task
Add PID and track variables for all charged final state particles and the invariant mass of the intermediate resonances to the ntuple. Also, add the standard variables from before for all particles in the decay chain, and include the kinematics both in the lab and the CMS frame.
Hint: Where to look
You need the variable collections and alias functions mentioned above! Take it one step at a time, it’s more lines of code than in the previous examples.
Hint: Partial solution for final state particles
This is how we add variables to the final state particles:
# variables for final states (electrons, positrons, pions) [S50]
fs_vars = vc.pid + vc.track + vc.track_hits + standard_vars
b_vars += vu.create_aliases_for_selected(
fs_vars,
"B0 -> [J/psi -> ^e+ ^e-] [K_S0 -> ^pi+ ^pi-]",
prefix=["ep", "em", "pip", "pim"],
) # [E50]
Next, do the same for the \(J/\Psi\) and the \(K_S^0\) in a similar fashion.
Hint: CMS variables
The bit about the CMS variables is tricky. First create a variable
cmskinematics with the create_aliases function. Use “CMS” as the
prefix. What do you need to specify for the wrapper?
Then use your cmskinematics variable together with
create_aliases_for_selected to add these variables to the relevant
particles.
Hint: Partial solution for the CMS variables
This is the code for the first part of the last hint:
# also add kinematic variables boosted to the center of mass frame (CMS) [S60]
# for all particles
cmskinematics = vu.create_aliases(
vc.kinematics, "useCMSFrame({variable})", "CMS"
) # [E60]
Solution
#!/usr/bin/env python3
import sys
import basf2 as b2
import modularAnalysis as ma
import stdV0s
import variables.collections as vc
import variables.utils as vu
# get input file number from the command line
filenumber = sys.argv[1]
# create path
main = b2.Path()
# load input data from mdst/udst file
ma.inputMdstList(
filelist=[b2.find_file(f"starterkit/2021/1111540100_eph3_BGx0_{filenumber}.root", "examples")],
path=main,
)
# fill final state particle lists
ma.fillParticleList(
"e+:uncorrected",
"electronID > 0.1 and dr < 0.5 and abs(dz) < 2 and thetaInCDCAcceptance",
path=main,
)
stdV0s.stdKshorts(path=main)
# combine final state particles to form composite particles
ma.reconstructDecay(
"J/psi:ee -> e+:uncorrected e-:uncorrected", cut="abs(dM) < 0.11", path=main
)
# combine J/psi and KS candidates to form B0 candidates
ma.reconstructDecay(
"B0 -> J/psi:ee K_S0:merged",
cut="Mbc > 5.2 and abs(deltaE) < 0.15",
path=main,
)
# match reconstructed with MC particles
ma.matchMCTruth("B0", path=main)
# create list of variables to save into the output file
b_vars = []
standard_vars = vc.kinematics + vc.mc_kinematics + vc.mc_truth
b_vars += vc.deltae_mbc
b_vars += standard_vars
# variables for final states (electrons, positrons, pions) [S50]
fs_vars = vc.pid + vc.track + vc.track_hits + standard_vars
b_vars += vu.create_aliases_for_selected(
fs_vars,
"B0 -> [J/psi -> ^e+ ^e-] [K_S0 -> ^pi+ ^pi-]",
prefix=["ep", "em", "pip", "pim"],
) # [E50]
# variables for J/Psi, KS
jpsi_ks_vars = vc.inv_mass + standard_vars
b_vars += vu.create_aliases_for_selected(jpsi_ks_vars, "B0 -> ^J/psi ^K_S0")
# also add kinematic variables boosted to the center of mass frame (CMS) [S60]
# for all particles
cmskinematics = vu.create_aliases(
vc.kinematics, "useCMSFrame({variable})", "CMS"
) # [E60]
b_vars += vu.create_aliases_for_selected(
cmskinematics, "^B0 -> [^J/psi -> ^e+ ^e-] [^K_S0 -> ^pi+ ^pi-]"
)
# save variables to an output file (ntuple)
ma.variablesToNtuple(
"B0",
variables=b_vars,
filename="Bd2JpsiKS.root",
treename="tree",
path=main,
)
# start the event loop (actually start processing things)
b2.process(main)
# print out the summary
print(b2.statistics)
Hint
To get more information about the aliases we are creating, simply use
VariableManager.printAliases (vm.printAliases()) just before
processing your path.
See also
Some more example steering files that center around the VariableManager
can be found on GitLab.
Exercise
Run your steering file one last time to check that it works!
Key points
The
modularAnalysismodule contains most of what you’ll need for nowinputMdstListis used to load datafillParticleListadds particles into a listreconstructDecaycombined FSPs from different lists to “reconstruct” particlesmatchMCTruthmatches MCvariablesToNtuplesaves an output fileDon’t forget
process(path)or nothing happens
Stuck? We can help!
If you get stuck or have any questions to the online book material, the #starterkit-workshop channel in our chat is full of nice people who will provide fast help.
Refer to Collaborative Tools. for other places to get help if you have specific or detailed questions about your own analysis.
Improving things!
If you know how to do it, we recommend you to report bugs and other requests
with GitLab.
Make sure to use the documentation-training label of the basf2 project.
If you just want to give very quick feedback, use the last box “Quick feedback”.
Please make sure to be as precise as possible to make it easier for us to fix things! So for example:
typos (where?)
missing bits of information (what?)
bugs (what did you do? what goes wrong?)
too hard exercises (which one?)
etc.
If you are familiar with git and want to create your first merge request for the software, take a look at How to contribute. We’d be happy to have you on the team!
Quick feedback!
Authors of this lesson
Frank Meier, Kilian Lieret (minor improvements)