3.7. Workflow Management#
Increasingly complex HEP analyses require many separate analysis steps on Monte Carlo simulations and data. In a typical Belle II analysis, we run skims, reconstructions and offline analysis on different computing resources:
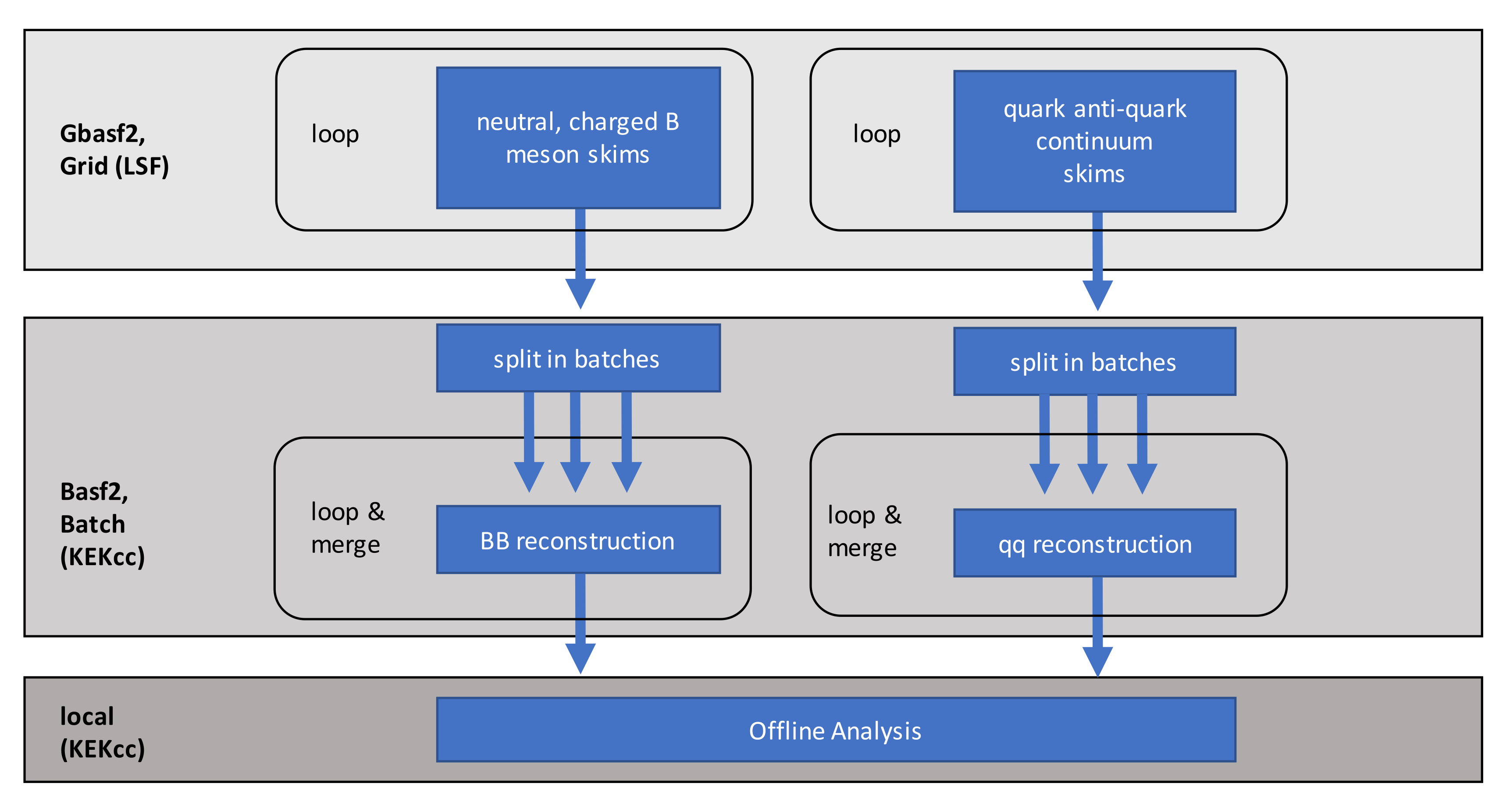
Fig. 3.36 Analysis steps on different computing resources in a sample Belle II analysis.#
The sequence of all processing steps required for your analysis is a workflow. In your own interest (as well as in the interest of analysts following after you) you should set up your analysis in a workflow management system, i.e. automatize the entire workflow execution.
Currently, there is a lack of documentation of interplay of the different scripts and jobs, which are executed manually one-by-one by the analyst. This is error-prone, time-consuming and deteriorates the reproducibility of results, the transparency of collaborative reviews and hinders data preservation efforts.
In so-called workflow management tools, dependencies between processing steps are made explicit in a stand-alone executable, including job submission to remote computing resources, parallel computing etc. Previous boilerplate code (such as custom bash scripts) becomes obsolete.
A workflow is visualized in a directed acyclic graph (DAG), which illustrates the dependencies between all processing steps. The DAG for a typical Belle II analysis quickly gets large, and workflow management tools can save you lots of headaches:
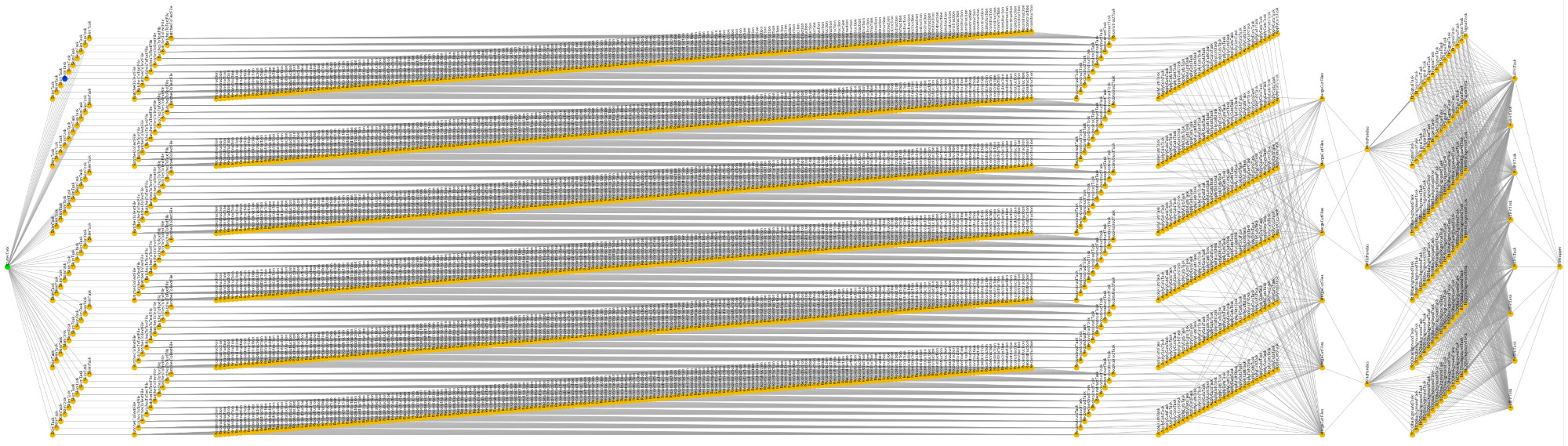
Fig. 3.37 Directed acyclic graph (DAG) for a sample Belle II analysis.#
A wide variety of workflow management tools exists (see for example here). For Belle II analyses, the b2luigi (based on the luigi framework) and snakemake workflow management tools are particulary useful (see e.g. our comparison). In general, each processing step is implemented as a task in the workflow, with its input(s) and output(s). A task is automatically scheduled for execution by the workflow management tool, as soon as all of its input(s) are existing but not all of its output(s). If output(s) to a task are already existing upon launch, the corresponding task will not be run again.
In this lesson, we build a minimalistic Belle II analysis in both tools, employing gbasf2, basf2 and the LSF batch system: