3.7.3. snakemake#
Snakemake is a workflow management framework designed for uses in research, with extensive support for environment management and remote execution. It features a very simple Python-based syntax and accommodates shell commands and external scripts in multiple programming languages with minimal adaptations. A full guide can be found elsewhere.
Let us reproduce the full Belle II workflow that we dicussed for b2luigi.
Again, we submit four skims (two for B mesons and two for quark-antiquark continuum) to the grid, which are each reconstructed in three batches on the KEKcc batch system. The reconstructed batches are merged and finally we fill some histograms in the offline analysis.
The directed acyclic graph (DAG) looks like this:
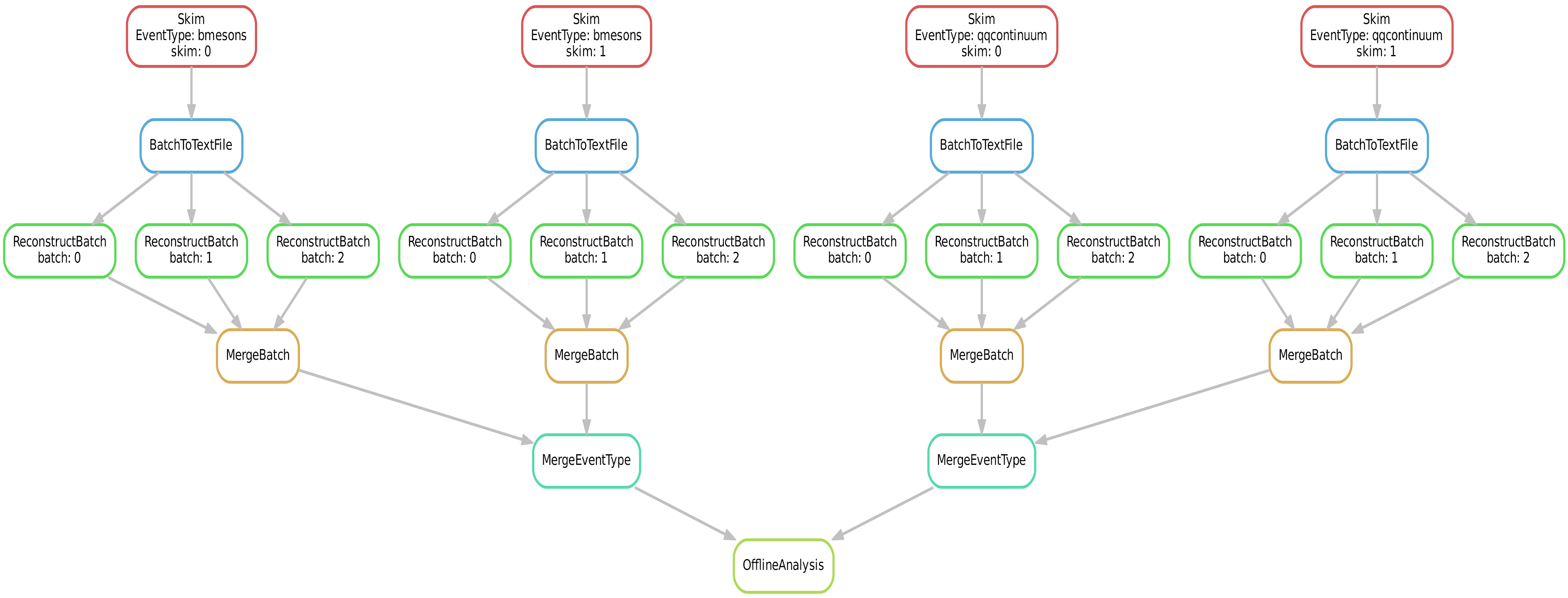
Now, let us look at the source code step by step:
Stuck? We can help!
If you get stuck or have any questions to the online book material, the #starterkit-workshop channel in our chat is full of nice people who will provide fast help.
Refer to Collaborative Tools. for other places to get help if you have specific or detailed questions about your own analysis.
Improving things!
If you know how to do it, we recommend you to report bugs and other requests
with GitLab.
Make sure to use the documentation-training label of the basf2 project.
If you just want to give us feedback, please open a GitLab issue and add the label online_book to it.
Please make sure to be as precise as possible to make it easier for us to fix things! So for example:
typos (where?)
missing bits of information (what?)
bugs (what did you do? what goes wrong?)
too hard exercises (which one?)
etc.
If you are familiar with git and want to create your first merge request for the software, take a look at How to contribute. We’d be happy to have you on the team!
Quick feedback!
Do you want to leave us some feedback? Open a GitLab issue and add the label online_book to it.