3.5.1. ROOT#
If you don’t know about ROOT yet, check out the section ROOT: a nano introduction. You can find the documentation for RDataFrames here.
RDataFrames#
RDataFrames are ROOT’s recommended interface to analysis. They can interpret trees in a root file into a table-like object.
import ROOT
The RDataFrame constructor takes the name of a tree and one or more
files.
df = ROOT.RDataFrame("treename", "file.root")
# or
df = ROOT.RDataFrame("treename", ["file1.root", "file2.root", ...])
The location of the files can be local or remote.
To follow this tutorial, download the ntuple from https://rebrand.ly/00vvyzg (it is the same as the one in the pandas tutorial) and read it:
filepath = "pandas_tutorial_ntuple.root"
df = ROOT.RDataFrame("b0phiKs", filepath)
Inspect the contents of a tree
df.Describe() # new in ROOT v6.26.00
Dataframe from TChain b0phiKs in file /path/to/pandas_tutorial_ntuple.root
Property Value
-------- -----
Columns in total 50
Columns from defines 0
Event loops run 0
Processing slots 1
Column Type Origin
------ ---- ------
B0_CosTBTO Float_t Dataset
B0_CosTBz Float_t Dataset
B0_ErrM Float_t Dataset
B0_K_S0_ErrM Float_t Dataset
B0_K_S0_M Float_t Dataset
B0_K_S0_SigM Float_t Dataset
B0_M Float_t Dataset
B0_R2 Float_t Dataset
B0_SigM Float_t Dataset
B0_ThrustB Float_t Dataset
...
Inspect the contents of one or more columns
df.Display(["B0_M", "B0_ErrM"], 5).Print()
+-----+----------+-------------+
| Row | B0_M | B0_ErrM |
+-----+----------+-------------+
| 0 | 5.02445f | 0.0224362f |
+-----+----------+-------------+
| 1 | 5.10793f | 0.0823563f |
+-----+----------+-------------+
| 2 | 5.11921f | 0.0868997f |
+-----+----------+-------------+
| 3 | 5.36136f | 0.00969569f |
+-----+----------+-------------+
| 4 | 5.30105f | 0.00664467f |
+-----+----------+-------------+
Get the number of events in this tree
df.Count().GetValue()
Note
RDataFrames are lazy which means that operations on them are not carried out immediately, but only
if a user requests it. For example, df.Count() does not return the number of events, but a Result
object that promises to compute the number of events in the future. The GetValue() method extracts the actual
result for us.
Functionality for data analysis#
Think of an RDataFrame as a table that you can use to compute new columns from existing ones and filter based on various conditions.
df_filtered = df.Filter("condition", "optional name for this cut")
The condition can be passed either as a C++ expression in a string or as a python function. Defining new columns works in the same way:
df_new = df_filtered.Define("columnname", "c++ expression")
Note
Filter and Define do not mutate the
dataframe object but rather return a new RDataFrame object. These operations are
also lazy meaning that nothing is computed until the result is
actually requested by the user.
For example, we could define two new columns in our RDataFrame like this:
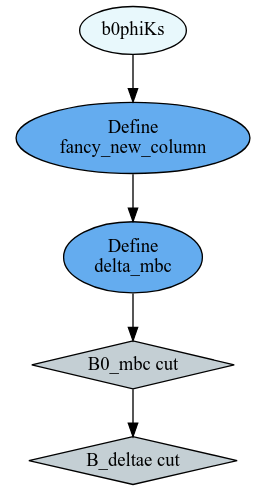
df = df.Define("fancy_new_column", "TMath::Power((B0_deltae * B0_et), 2) / TMath::Sin(B0_cc2)")\
.Define("delta_mbc", "B0_M - B0_mbc")
and filter it like this:
df = df.Filter("B0_mbc>5.2", "B0_mbc cut")\
.Filter("B0_deltae>-1", "B_deltae cut")
Because of RDataFrame’s laziness, these operations return almost instantly. The computations are only “booked”.
Exercise
Create two RDataFrames, one for Signal and one for Background only.
Hint
Split between signal and background using the B0_isSignal column.
Solution
bkgd_df = df.Filter("B0_isSignal==0")
signal_df = df.Filter("B0_isSignal==1")
Experimental new feature: Systematic variations#
RDataFrames offer a declarative way to define systematic variations of columns:
nominal_df = df.Vary("pt", "ROOT::RVecD{pt*0.9, pt*1.1}", ["down", "up"])
.Define(...)
.Filter(...)
histo = ROOT.RDF.Experimental.VariationsFor(nominal_df)
histo["nominal"].Draw()
histo["pt:down"].Draw("SAME")
Interoperability#
The columns in RDataFrames can be converted to numpy arrays for usecases where you don’t want to continue working with ROOT.
Converting to numpy is one example of the user requesting to get the data and therefore triggering the execution of all previously booked computations. You can convert one or more columns at a time:
delta_mbc = df.AsNumpy(["delta_mbc"])
We get back a dict
{'delta_mbc': ndarray([-0.18043327, -0.10750389, -0.09657669, ..., 0.02187395,
0.04272509, 0.01566553], dtype=float32)}
and can now continue to work on the result outside of the ROOT-world.
Inspection#
RDataFrames offer easily accessible methods to track down what actually happened in a computation.
For example get a report of the efficiencies of each filter applied:
df.Report().Print()
B0_mbc cut: pass=327351 all=329135 -- eff=99.46 % cumulative eff=99.46 %
B_deltae cut: pass=327349 all=327351 -- eff=100.00 % cumulative eff=99.46 %
Or get the computational graph
# visualize the computation graph
ROOT.RDF.SaveGraph(df, "DAG.dot")
from graphviz import Source
Source.from_file("DAG.dot")
Scaling up#
RDataFrames have the (as of now still experimental) option to run distributed on a cluster (eg. dask) to scale up your analysis.
import dask_jobqueue
from dask.distributed import Client
import ROOT
DistRDataFrame = ROOT.RDF.Experimental.Distributed.Dask.RDataFrame
cluster = dask_jobqueue.SLURMCluster(
name="myanalysis",
cores=1,
queue="my-slurm-cluster",
memory="4GB",
job_extra_directives=[...],
)
cluster.scale(90)
client = Client(cluster)
df = DistRDataFrame("treename", filelist, daskclient=client)
Note that the interface to distributed RDataFrames is the same as normal RDataFrames, so there’s no need to change any analysis code.
Dask comes with a handy dashboard that shows the progress of all tasks across the workers, a flamegraph and many more monitoring utilities.